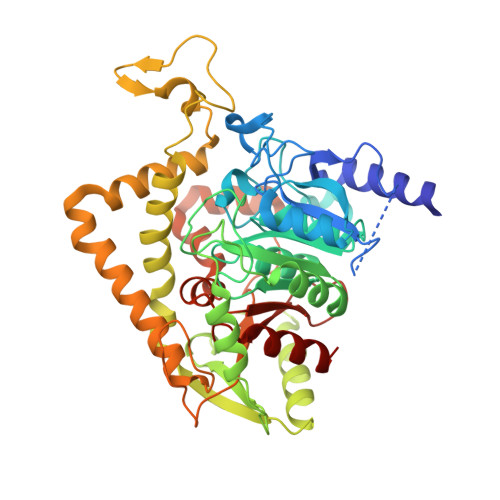
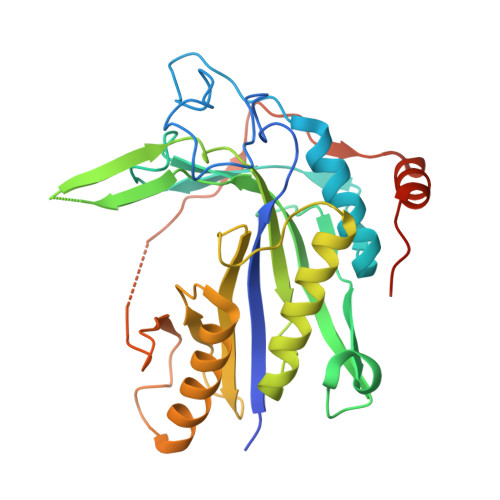
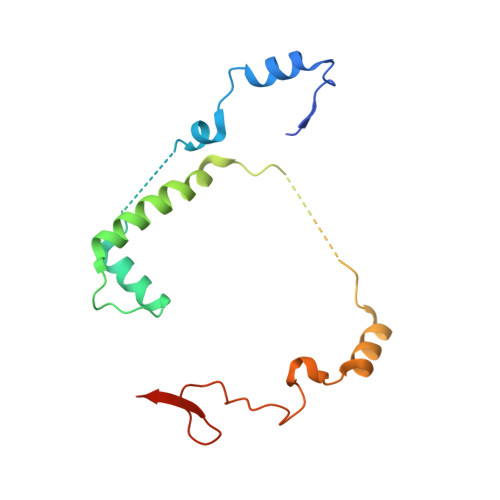
Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Araiso, Y., Huot, J.L., Sekiguchi, T., Frechin, M., Fischer, F., Enkler, L., Senger, B., Ishitani, R., Becker, H.D., Nureki, O.(2014) Nucleic Acids Res 42: 6052-6063
- PubMed: 24692665
- DOI: https://doi.org/10.1093/nar/gku234
- Primary Citation of Related Structures:
4N0H, 4N0I - PubMed Abstract:
Yeast mitochondrial Gln-mtRNAGln is synthesized by the transamidation of mischarged Glu-mtRNAGln by a non-canonical heterotrimeric tRNA-dependent amidotransferase (AdT). The GatA and GatB subunits of the yeast AdT (GatFAB) are well conserved among bacteria and eukaryota, but the GatF subunit is a fungi-specific ortholog of the GatC subunit found in all other known heterotrimeric AdTs (GatCAB). Here we report the crystal structure of yeast mitochondrial GatFAB at 2.0 Å resolution. The C-terminal region of GatF encircles the GatA-GatB interface in the same manner as GatC, but the N-terminal extension domain (NTD) of GatF forms several additional hydrophobic and hydrophilic interactions with GatA. NTD-deletion mutants displayed growth defects, but retained the ability to respire. Truncation of the NTD in purified mutants reduced glutaminase and transamidase activities when glutamine was used as the ammonia donor, but increased transamidase activity relative to the full-length enzyme when the donor was ammonium chloride. Our structure-based functional analyses suggest the NTD is a trans-acting scaffolding peptide for the GatA glutaminase active site. The positive surface charge and novel fold of the GatF-GatA interface, shown in this first crystal structure of an organellar AdT, stand in contrast with the more conventional, negatively charged bacterial AdTs described previously.
Organizational Affiliation:
Unité Mixte de Recherche 7156 Génétique Moléculaire Génomique Microbiologie, Centre National de la Recherche Scientifique, Université de Strasbourg, F-67084 Strasbourg, France.