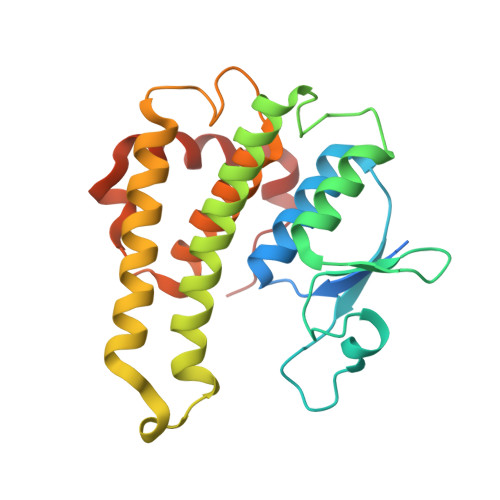
Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h)
Toro, R., Bhosle, R., Vetting, M.W., Al Obaidi, N.F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Stead, M., Washington, E., Scott Glenn, A., Chowdhury, S., Evans, B., Hillerich, B., Love, J., Seidel, R.D., Imker, H.J., Gerlt, J.A., Armstrong, R.N., Almo, S.C.To be published.