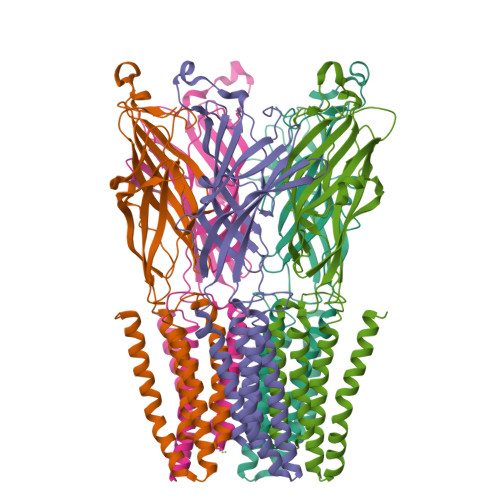
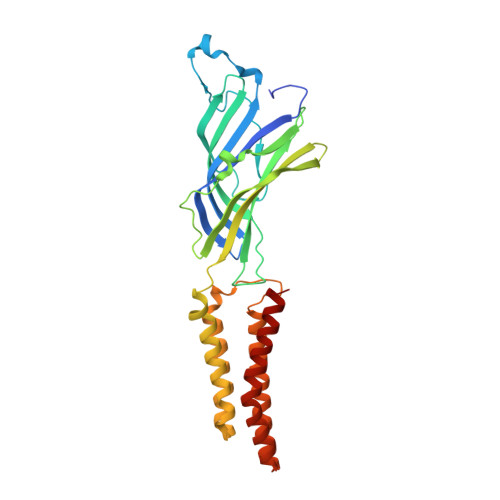
Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography.
Gonzalez-Gutierrez, G., Cuello, L.G., Nair, S.K., Grosman, C.(2013) Proc Natl Acad Sci U S A 110: 18716-18721
- PubMed: 24167270
- DOI: https://doi.org/10.1073/pnas.1313156110
- Primary Citation of Related Structures:
4LMJ, 4LMK, 4LML - PubMed Abstract:
Cryoelectron microscopy and X-ray crystallography have recently been used to generate structural models that likely represent the unliganded closed-channel conformation and the fully liganded open-channel conformation of different members of the nicotinic-receptor superfamily. To characterize the structure of the closed-channel conformation in its liganded state, we identified a number of positions in the loop between transmembrane segments 2 (M2) and 3 (M3) of a proton-gated ortholog from the bacterium Gloeobacter violaceus (GLIC) where mutations to alanine reduce the liganded-gating equilibrium constant, and solved the crystal structures of two such mutants (T25'A and Y27'A) at pH ~4.0. At the level of backbone atoms, the liganded closed-channel model presented here differs from the liganded open-channel structure of GLIC in the pre-M1 linker, the M3-M4 loop, and much more prominently, in the extracellular half of the pore lining, where the more pronounced tilt of the closed-channel M2 α-helices toward the pore's long axis narrows the permeation pathway. On the other hand, no differences between the liganded closed-channel and open-channel models could be detected at the level of the extracellular domain, where conformational changes are expected to underlie the low-to-high proton-affinity switch that drives gating of proton-bound channels. Thus, the liganded closed-channel model is nearly indistinguishable from the recently described "locally closed" structure. However, because cross-linking strategies (which could have stabilized unstable conformations) and mutations involving ionizable side chains (which could have affected proton-gated channel activation) were purposely avoided, we favor the notion that this structure represents one of the end states of liganded gating rather than an unstable intermediate.
Organizational Affiliation:
Department of Molecular and Integrative Physiology, Center for Biophysics and Computational Biology, Department of Biochemistry, Institute for Genomic Biology, and Neuroscience Program, University of Illinois at Urbana-Champaign, Urbana, IL 61801.