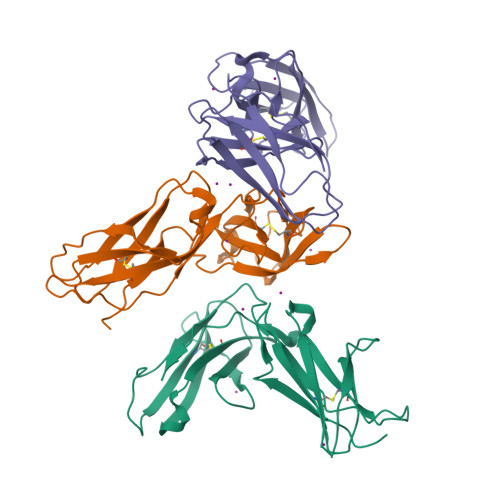
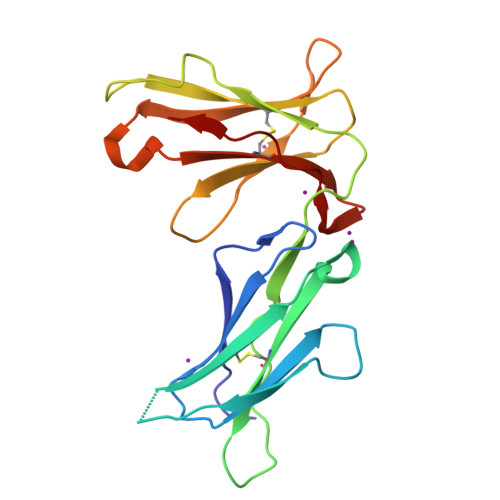
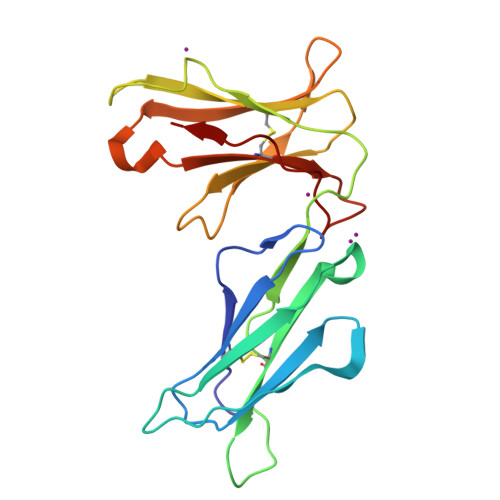
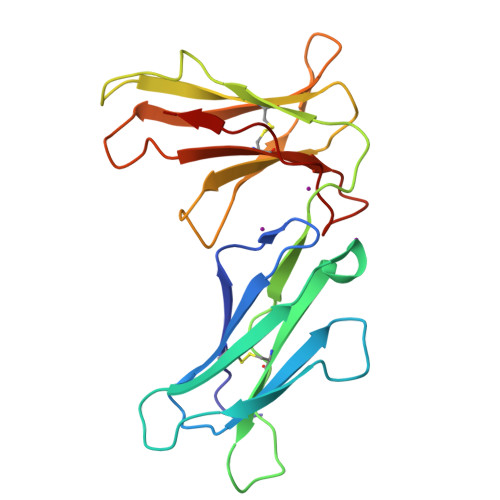
Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Nam, G., Shi, Y., Ryu, M., Wang, Q., Song, H., Liu, J., Yan, J., Qi, J., Gao, G.F.(2013) Protein Cell 4: 761-770
- PubMed: 23955630
- DOI: https://doi.org/10.1007/s13238-013-3908-x
- Primary Citation of Related Structures:
4LL9, 4LLA - PubMed Abstract:
Leukocyte immunoglobulin-like receptors (LILRs), also called CD85s, ILTs, or LIRs, are important mediators of immune activation and tolerance that contain tandem immunoglobulin (Ig)-like folds. There are 11 (in addition to two pseudogenes) LILRs in total, two with two Ig-like domains (D1D2) and the remaining nine with four Ig-like domains (D1D2D3D4). Thus far, the structural features of the D1D2 domains of LILR proteins are well defined, but no structures for the D3D4 domains have been reported. This is a very important field to be studied as it relates to the unknown functions of the D3D4 domains, as well as their relative orientation to the D1D2 domains on the cell surface. Here, we report the crystal structures of the D3D4 domains of both LILRB1 and LILRB2. The two Ig-like domains of both LILRB1-D3D4 and LILRB2-D3D4 are arranged at an acute angle (∼60°) to form a bent structure, resembling the structures of natural killer inhibitory receptors. Based on these two D3D4 domain structures and previously reported D1D2/HLA I complex structures, two alternative models of full-length (four Ig-like domains) LILR molecules bound to HLA I are proposed.
Organizational Affiliation:
CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, 100101, China.