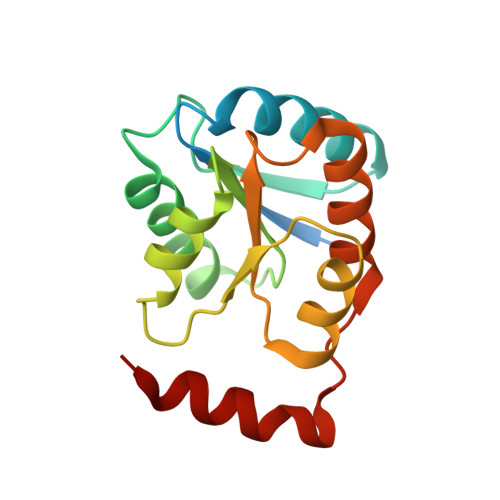
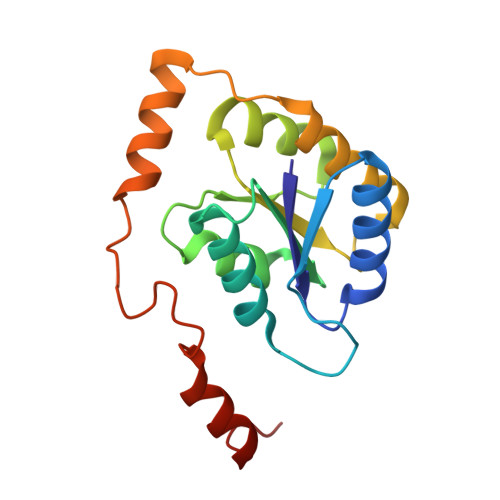
Crystal structure and substrate specificity of D-galactose-6-phosphate isomerase complexed with substrates.
Jung, W.S., Singh, R.K., Lee, J.K., Pan, C.H.(2013) PLoS One 8: e72902-e72902
- PubMed: 24015281
- DOI: https://doi.org/10.1371/journal.pone.0072902
- Primary Citation of Related Structures:
4LFK, 4LFL, 4LFM, 4LFN - PubMed Abstract:
D-Galactose-6-phosphate isomerase from Lactobacillus rhamnosus (LacAB; EC 5.3.1.26), which is encoded by the tagatose-6-phosphate pathway gene cluster (lacABCD), catalyzes the isomerization of D-galactose-6-phosphate to D-tagatose-6-phosphate during lactose catabolism and is used to produce rare sugars as low-calorie natural sweeteners. The crystal structures of LacAB and its complex with D-tagatose-6-phosphate revealed that LacAB is a homotetramer of LacA and LacB subunits, with a structure similar to that of ribose-5-phosphate isomerase (Rpi). Structurally, LacAB belongs to the RpiB/LacAB superfamily, having a Rossmann-like αβα sandwich fold as has been identified in pentose phosphate isomerase and hexose phosphate isomerase. In contrast to other family members, the LacB subunit also has a unique α7 helix in its C-terminus. One active site is distinctly located at the interface between LacA and LacB, whereas two active sites are present in RpiB. In the structure of the product complex, the phosphate group of D-tagatose-6-phosphate is bound to three arginine residues, including Arg-39, producing a different substrate orientation than that in RpiB, where the substrate binds at Asp-43. Due to the proximity of the Arg-134 residue and backbone Cα of the α6 helix in LacA to the last Asp-172 residue of LacB with a hydrogen bond, a six-carbon sugar-phosphate can bind in the larger pocket of LacAB, compared with RpiB. His-96 in the active site is important for ring opening and substrate orientation, and Cys-65 is essential for the isomerization activity of the enzyme. Two rare sugar substrates, D-psicose and D-ribulose, show optimal binding in the LacAB-substrate complex. These findings were supported by the results of LacA activity assays.
- Functional Food Center, Korea Institute of Science and Technology Gangneung Institute, Gangneung, Korea.
Organizational Affiliation: