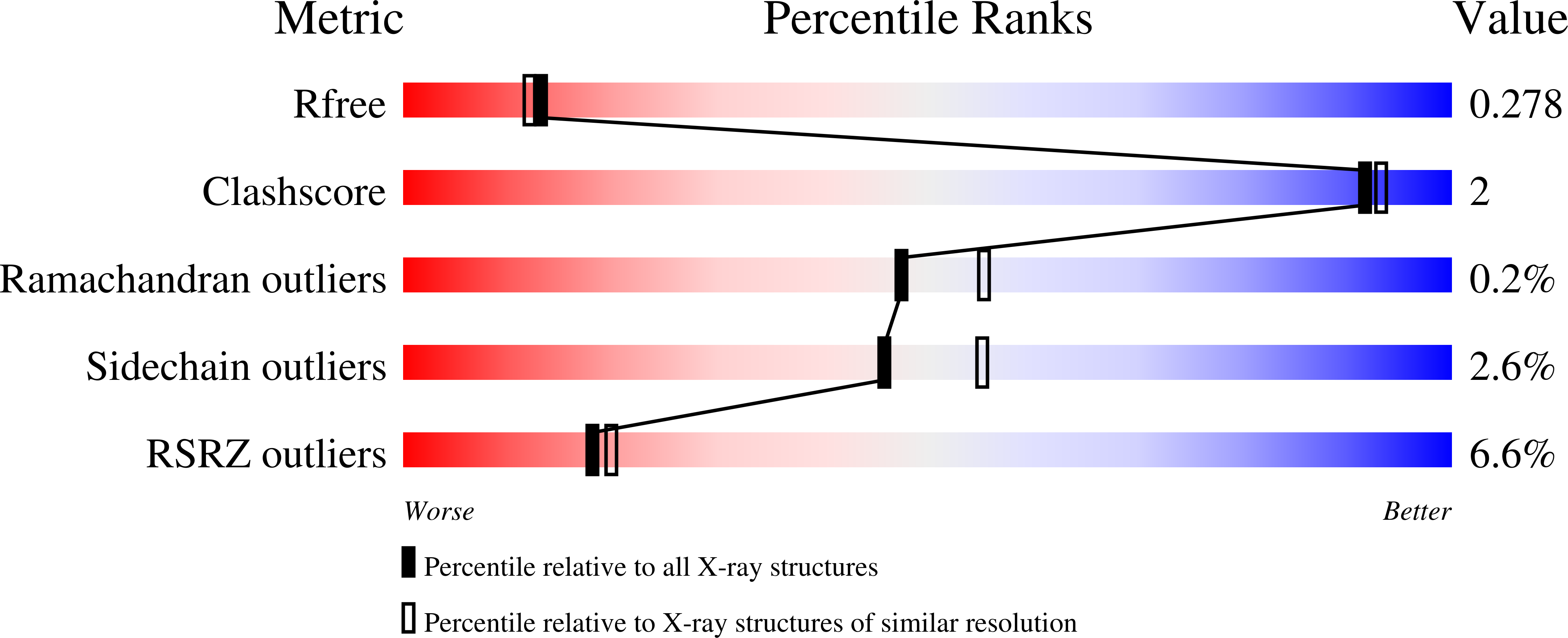
Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Jnoff, E., Albrecht, C., Barker, J.J., Barker, O., Beaumont, E., Bromidge, S., Brookfield, F., Brooks, M., Bubert, C., Ceska, T., Corden, V., Dawson, G., Duclos, S., Fryatt, T., Genicot, C., Jigorel, E., Kwong, J., Maghames, R., Mushi, I., Pike, R., Sands, Z.A., Smith, M.A., Stimson, C.C., Courade, J.P.(2014) ChemMedChem 9: 699-705
- PubMed: 24504667
- DOI: https://doi.org/10.1002/cmdc.201300525
- Primary Citation of Related Structures:
4L7B, 4L7C, 4L7D, 4N1B - PubMed Abstract:
An X-ray crystal structure of Kelch-like ECH-associated protein (Keap1) co-crystallised with (1S,2R)-2-[(1S)-1-[(1,3-dioxo-2,3-dihydro-1H-isoindol-2-yl)methyl]-1,2,3,4-tetrahydroisoquinolin-2-carbonyl]cyclohexane-1-carboxylic acid (compound (S,R,S)-1 a) was obtained. This X-ray crystal structure provides breakthrough experimental evidence for the true binding mode of the hit compound (S,R,S)-1 a, as the ligand orientation was found to differ from that of the initial docking model, which was available at the start of the project. Crystallographic elucidation of this binding mode helped to focus and drive the drug design process more effectively and efficiently.
Organizational Affiliation:
UCB Pharma, UCB NewMedicines, Chemin du Foriest, 1420 Braine-l'Alleud (Belgium). eric.jnoff@ucb.com.