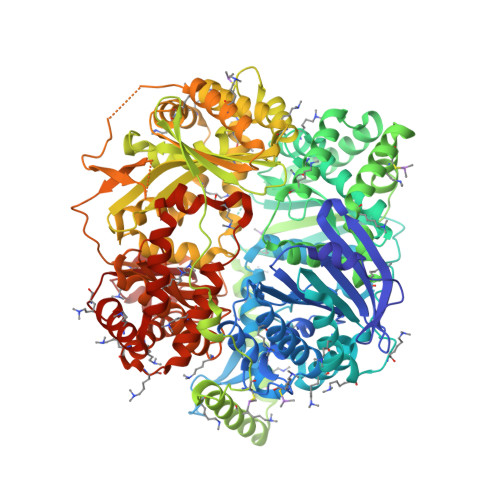
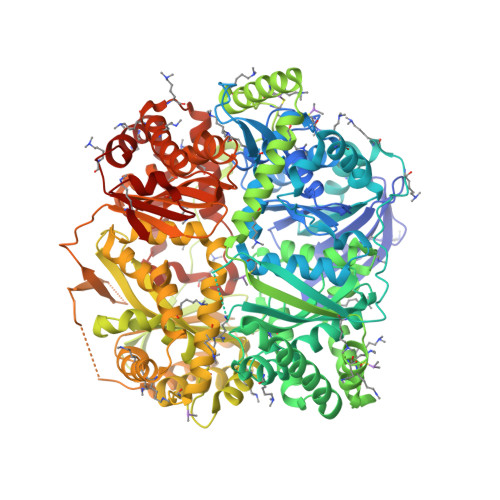
Molecular basis of substrate recognition and degradation by human presequence protease.
King, J.V., Liang, W.G., Scherpelz, K.P., Schilling, A.B., Meredith, S.C., Tang, W.J.(2014) Structure 22: 996-1007
- PubMed: 24931469
- DOI: https://doi.org/10.1016/j.str.2014.05.003
- Primary Citation of Related Structures:
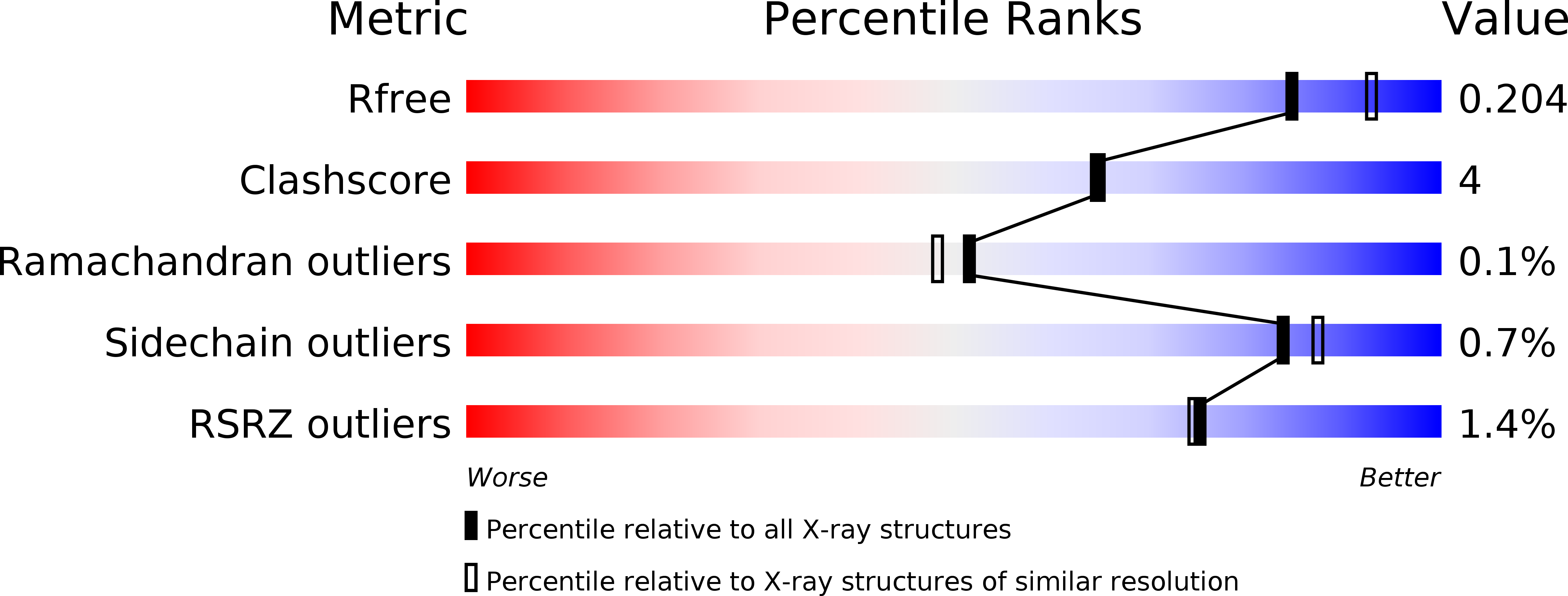
4L3T, 4NGE - PubMed Abstract:
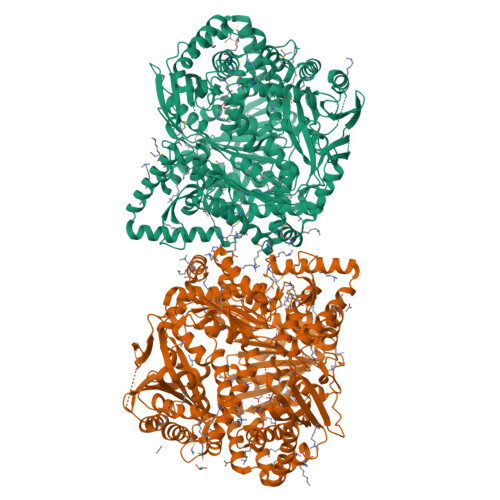
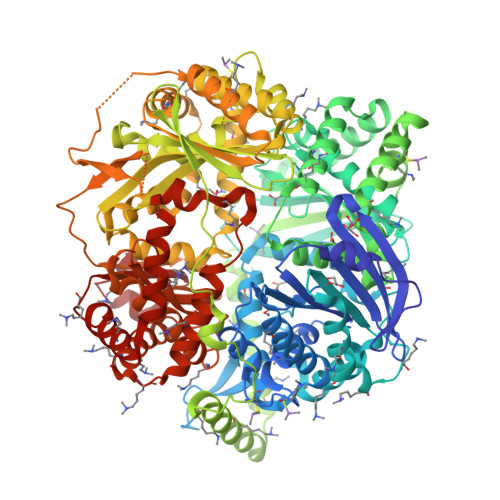
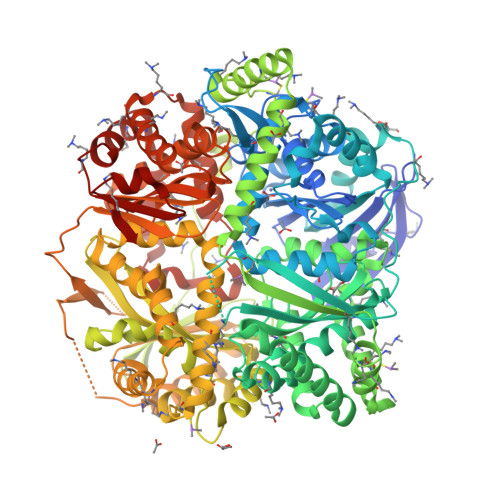
Human presequence protease (hPreP) is an M16 metalloprotease localized in mitochondria. There, hPreP facilitates proteostasis by utilizing an ∼13,300-Å(3) catalytic chamber to degrade a diverse array of potentially toxic peptides, including mitochondrial presequences and β-amyloid (Aβ), the latter of which contributes to Alzheimer disease pathogenesis. Here, we report crystal structures for hPreP alone and in complex with Aβ, which show that hPreP uses size exclusion and charge complementation for substrate recognition. These structures also reveal hPreP-specific features that permit a diverse array of peptides, with distinct distributions of charged and hydrophobic residues, to be specifically captured, cleaved, and have their amyloidogenic features destroyed. SAXS analysis demonstrates that hPreP in solution exists in dynamic equilibrium between closed and open states, with the former being preferred. Furthermore, Aβ binding induces the closed state and hPreP dimerization. Together, these data reveal the molecular basis for flexible yet specific substrate recognition and degradation by hPreP.
Organizational Affiliation:
Ben May Department for Cancer Research, The University of Chicago, 929 E. 57(th) Street, Chicago, IL 60637, USA.