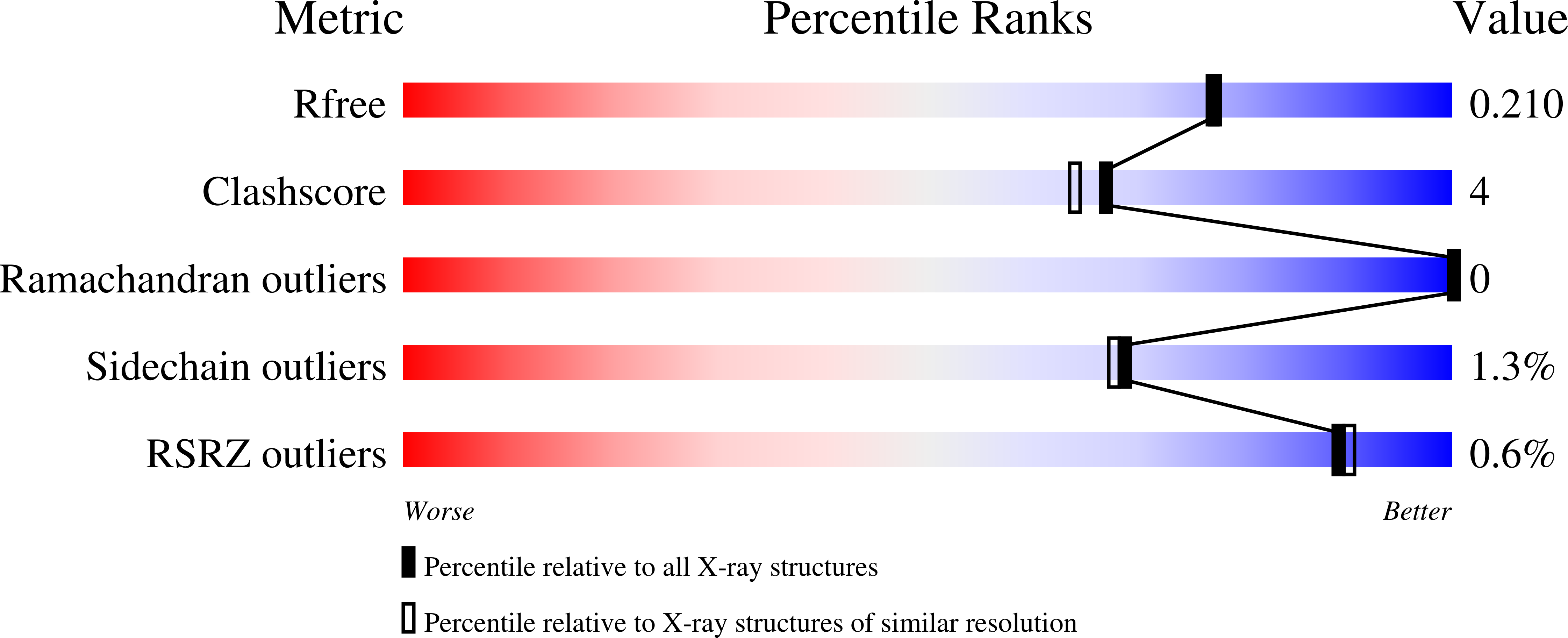
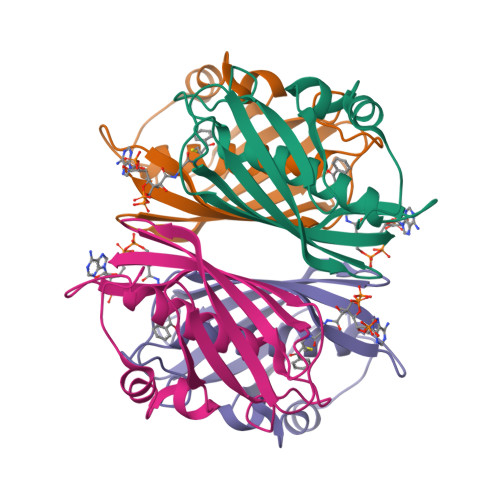
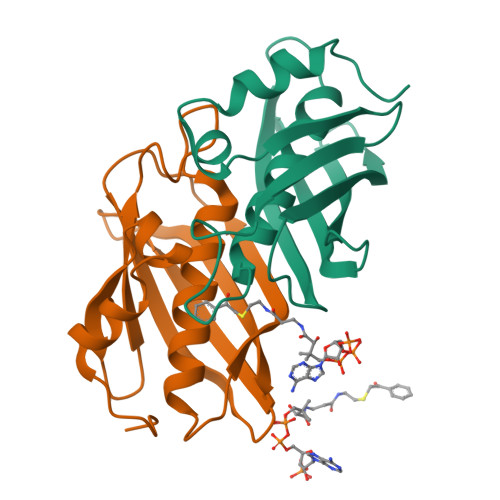
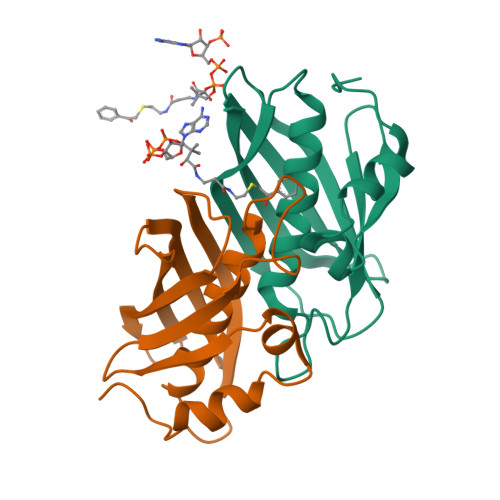
Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Wu, R., Latham, J.A., Chen, D., Farelli, J., Zhao, H., Matthews, K., Allen, K.N., Dunaway-Mariano, D.(2014) Biochemistry 53: 4788-4805
- PubMed: 25010423
- DOI: https://doi.org/10.1021/bi500334v
- Primary Citation of Related Structures:
4K49, 4K4A, 4K4B, 4K4C, 4K4D - PubMed Abstract:
Herein, the structural determinants for substrate recognition and catalysis in two hotdog-fold thioesterase paralogs, YbdB and YdiI from Escherichia coli, are identified and analyzed to provide insight into the evolution of biological function in the hotdog-fold enzyme superfamily. The X-ray crystal structures of YbdB and YdiI, in complex with inert substrate analogs, determined in this study revealed the locations of the respective thioester substrate binding sites and the identity of the residues positioned for substrate binding and catalysis. The importance of each of these residues was assessed through amino acid replacements followed by steady-state kinetic analyses of the corresponding site-directed mutants. Transient kinetic and solvent (18)O-labeling studies were then carried out to provide insight into the role of Glu63 posited to function as the nucleophile or general base in catalysis. Finally, the structure-function-mechanism profiles of the two paralogs, along with that of a more distant homolog, were compared to identify conserved elements of substrate recognition and catalysis, which define the core traits of the hotdog-fold thioesterase family, as well as structural features that are unique to each thioesterase. Founded on the insight gained from this analysis, we conclude that the promiscuity revealed by in vitro substrate activity determinations, and posited to facilitate the evolution of new biological function, is the product of intrinsic plasticity in substrate binding as well as in the catalytic mechanism.
Organizational Affiliation:
Department of Chemistry, Boston University , Boston, Massachusetts 02215, United States.