Disarming Burkholderia pseudomallei: Structural and Functional Characterization of a Disulfide Oxidoreductase (DsbA) Required for Virulence In Vivo.
Ireland, P.M., McMahon, R.M., Marshall, L.E., Halili, M., Furlong, E., Tay, S., Martin, J.L., Sarkar-Tyson, M.(2014) Antioxid Redox Signal 20: 606-617
- PubMed: 23901809
- DOI: https://doi.org/10.1089/ars.2013.5375
- Primary Citation of Related Structures:
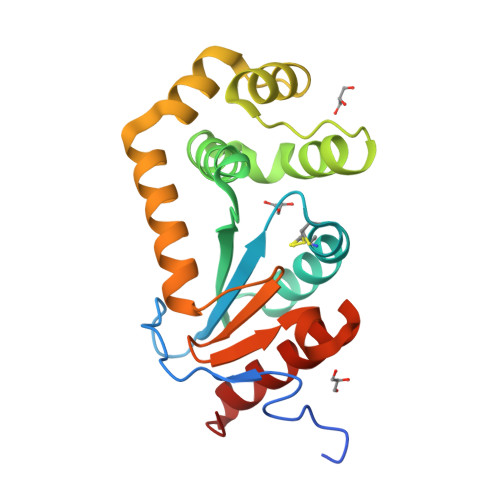
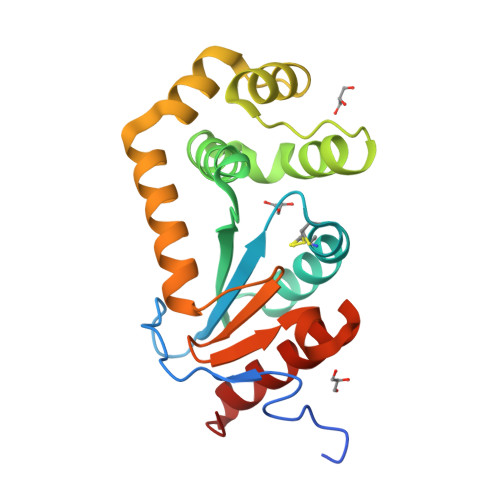
4K2D - PubMed Abstract:
The intracellular pathogen Burkholderia pseudomallei causes the disease melioidosis, a major source of morbidity and mortality in southeast Asia and northern Australia. The need to develop novel antimicrobials is compounded by the absence of a licensed vaccine and the bacterium's resistance to multiple antibiotics. In a number of clinically relevant Gram-negative pathogens, DsbA is the primary disulfide oxidoreductase responsible for catalyzing the formation of disulfide bonds in secreted and membrane-associated proteins. In this study, a putative B. pseudomallei dsbA gene was evaluated functionally and structurally and its contribution to infection assessed. Biochemical studies confirmed the dsbA gene encodes a protein disulfide oxidoreductase. A dsbA deletion strain of B. pseudomallei was attenuated in both macrophages and a BALB/c mouse model of infection and displayed pleiotropic phenotypes that included defects in both secretion and motility. The 1.9 Å resolution crystal structure of BpsDsbA revealed differences from the classic member of this family Escherichia coli DsbA, in particular within the region surrounding the active site disulfide where EcDsbA engages with its partner protein E. coli DsbB, indicating that the interaction of BpsDsbA with its proposed partner BpsDsbB may be distinct from that of EcDsbA-EcDsbB. This study has characterized BpsDsbA biochemically and structurally and determined that it is required for virulence of B. pseudomallei. These data establish a critical role for BpsDsbA in B. pseudomallei infection, which in combination with our structural characterization of BpsDsbA will facilitate the future development of rationally designed inhibitors against this drug-resistant organism.
Organizational Affiliation:
1 Defence Science and Technology Laboratory , Porton Down, Salisbury, Wiltshire, United Kingdom .