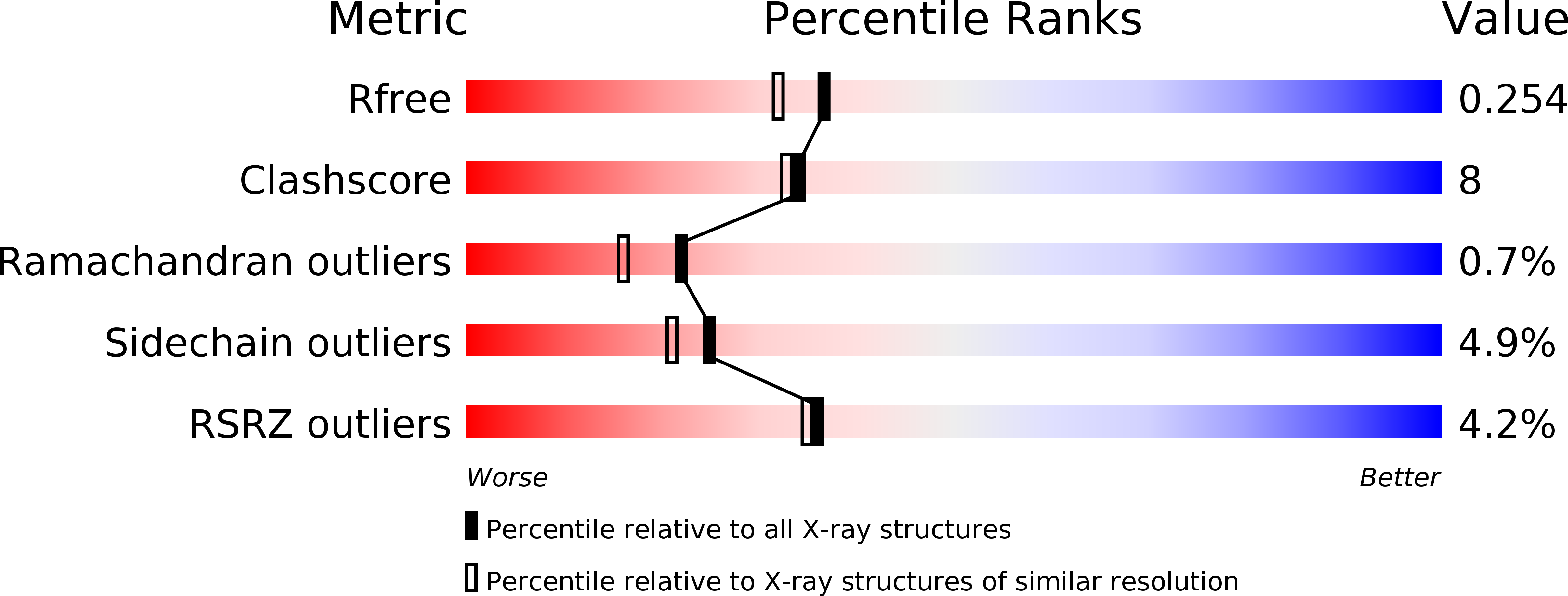
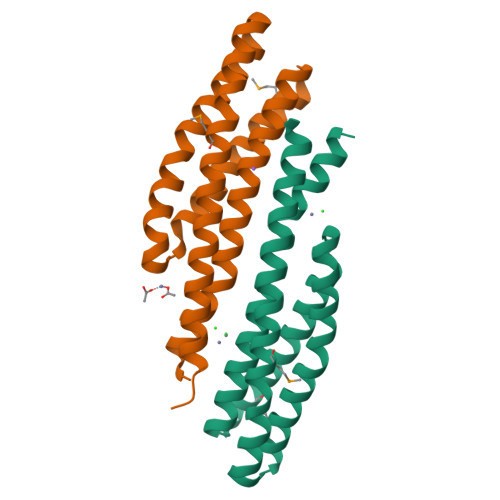
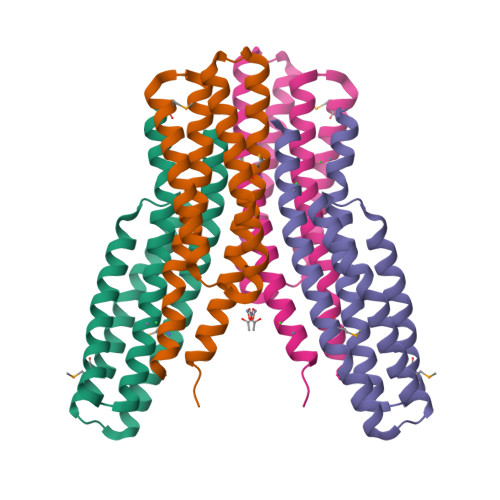
Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Pokkuluri, P.R., Dwulit-Smith, J., Duke, N.E., Wilton, R., Mack, J.C., Bearden, J., Rakowski, E., Babnigg, G., Szurmant, H., Joachimiak, A., Schiffer, M.(2013) Microbiologyopen 2: 766-777
- PubMed: 23897711
- DOI: https://doi.org/10.1002/mbo3.112
- Primary Citation of Related Structures:
4K08, 4K0D - PubMed Abstract:
Anaeromyxobacter dehalogenans is a δ-proteobacterium found in diverse soils and sediments. It is of interest in bioremediation efforts due to its dechlorination and metal-reducing capabilities. To gain an understanding on A. dehalogenans' abilities to adapt to diverse environments we analyzed its signal transduction proteins. The A. dehalogenans genome codes for a large number of sensor histidine kinases (HK) and methyl-accepting chemotaxis proteins (MCP); among these 23 HK and 11 MCP proteins have a sensor domain in the periplasm. These proteins most likely contribute to adaptation to the organism's surroundings. We predicted their three-dimensional folds and determined the structures of two of the periplasmic sensor domains by X-ray diffraction. Most of the domains are predicted to have either PAS-like or helical bundle structures, with two predicted to have solute-binding protein fold, and another predicted to have a 6-phosphogluconolactonase like fold. Atomic structures of two sensor domains confirmed the respective fold predictions. The Adeh_2942 sensor (HK) was found to have a helical bundle structure, and the Adeh_3718 sensor (MCP) has a PAS-like structure. Interestingly, the Adeh_3718 sensor has an acetate moiety bound in a binding site typical for PAS-like domains. Future work is needed to determine whether Adeh_3718 is involved in acetate sensing by A. dehalogenans.
Organizational Affiliation:
Biosciences, Argonne National Laboratory, 9700 South Cass Avenue, Lemont, Illinois, 60439.