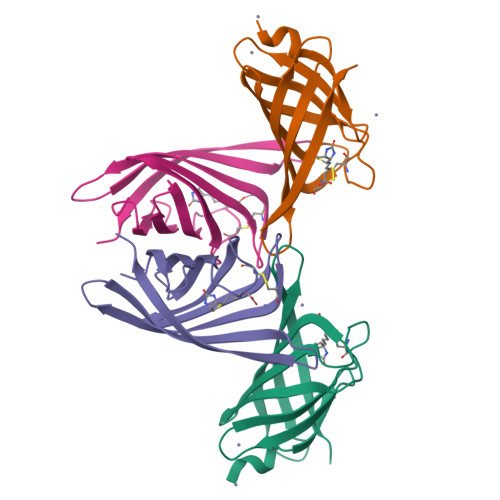
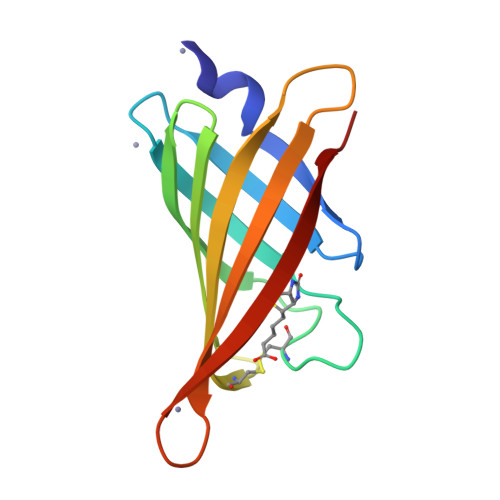
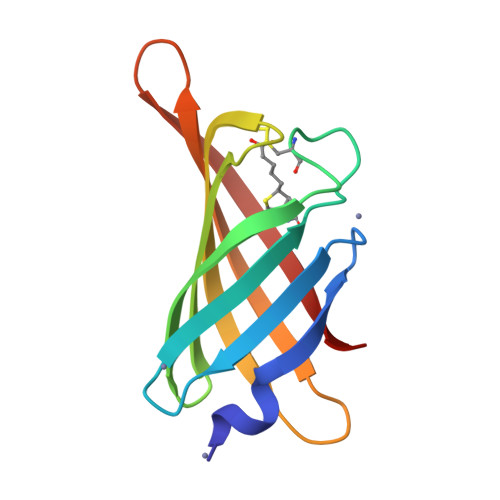
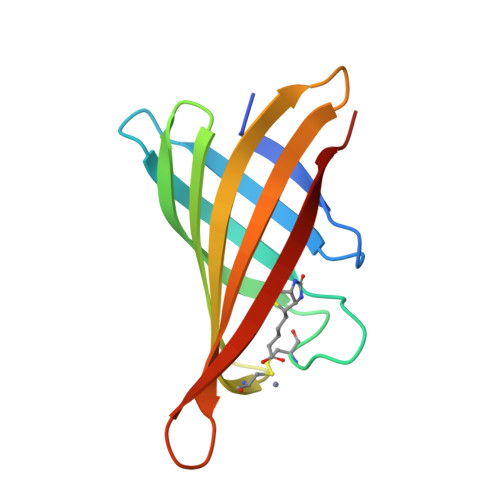
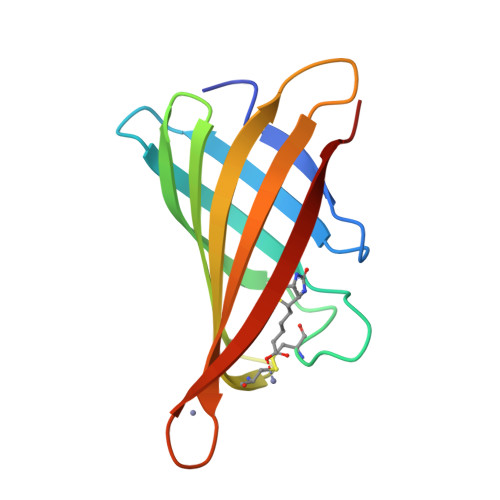
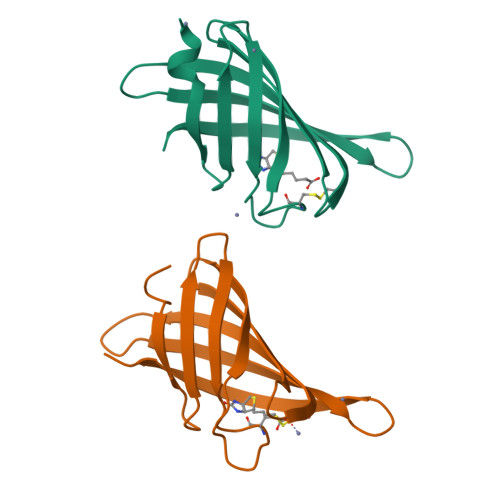
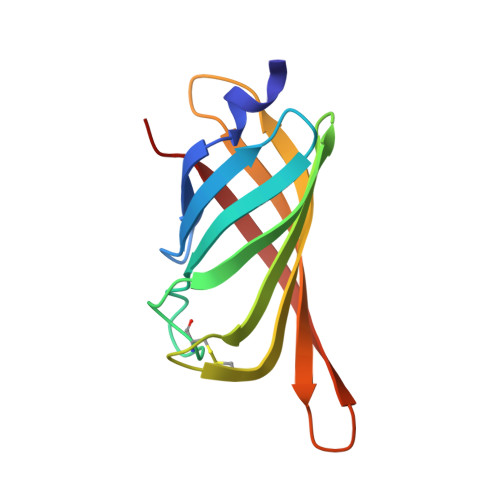
Structure-based engineering of streptavidin monomer with a reduced biotin dissociation rate.
Demonte, D., Drake, E.J., Lim, K.H., Gulick, A.M., Park, S.(2013) Proteins 81: 1621-1633
- PubMed: 23670729
- DOI: https://doi.org/10.1002/prot.24320
- Primary Citation of Related Structures:
4JNJ - PubMed Abstract:
We recently reported the engineering of monomeric streptavidin, mSA, corresponding to one subunit of wild type (wt) streptavidin tetramer. The monomer was designed by homology modeling, in which the streptavidin and rhizavidin sequences were combined to engineer a high affinity binding pocket containing residues from a single subunit only. Although mSA is stable and binds biotin with nanomolar affinity, its fast off rate (koff ) creates practical challenges during applications. We obtained a 1.9 Å crystal structure of mSA bound to biotin to understand their interaction in detail, and used the structure to introduce targeted mutations to improve its binding kinetics. To this end, we compared mSA to shwanavidin, which contains a hydrophobic lid containing F43 in the binding pocket and binds biotin tightly. However, the T48F mutation in mSA, which introduces a comparable hydrophobic lid, only resulted in a modest 20-40% improvement in the measured koff . On the other hand, introducing the S25H mutation near the bicyclic ring of bound biotin increased the dissociation half life (t½ ) from 11 to 83 min at 20°C. Molecular dynamics (MD) simulations suggest that H25 stabilizes the binding loop L3,4 by interacting with A47, and protects key intermolecular hydrogen bonds by limiting solvent entry into the binding pocket. Concurrent T48F or T48W mutation clashes with H25 and partially abrogates the beneficial effects of H25. Taken together, this study suggests that stabilization of the binding loop and solvation of the binding pocket are important determinants of the dissociation kinetics in mSA.
Organizational Affiliation:
Department of Chemical and Biological Engineering, University at Buffalo, Buffalo, New York, 14260.