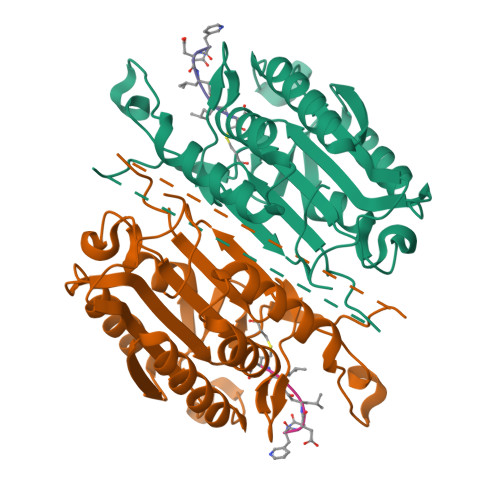
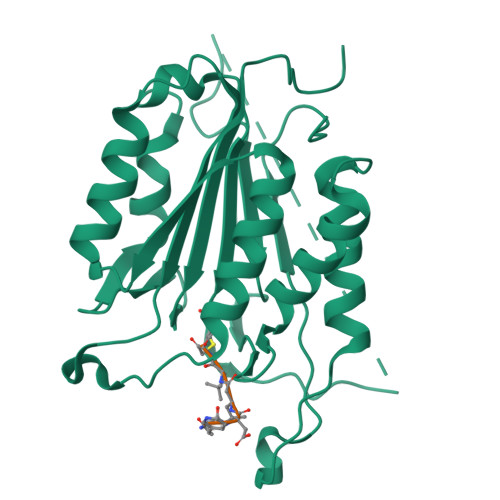
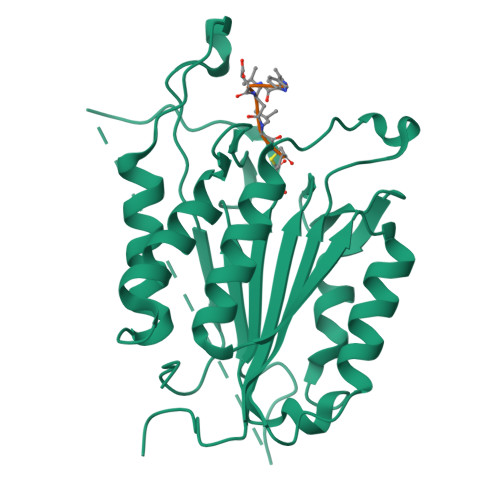
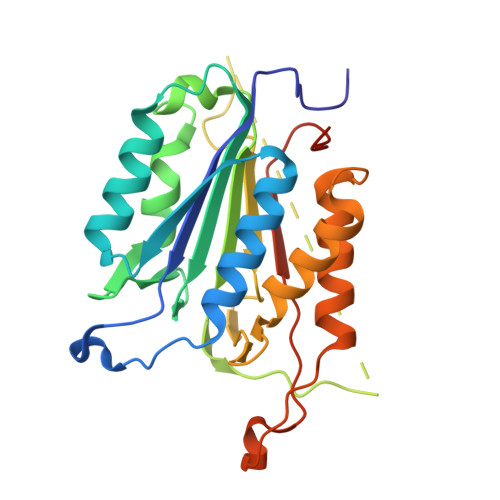
Selective Detection of Caspase-3 versus Caspase-7 Using Activity-Based Probes with Key Unnatural Amino Acids.
Vickers, C.J., Gonzalez-Paez, G.E., Wolan, D.W.(2013) ACS Chem Biol 8: 1558-1566
- PubMed: 23614665
- DOI: https://doi.org/10.1021/cb400209w
- Primary Citation of Related Structures:
4JJ7, 4JJ8, 4JJE - PubMed Abstract:
Caspases are required for essential biological functions, most notably apoptosis and pyroptosis, but also cytokine production, cell proliferation, and differentiation. One of the most well studied members of this cysteine protease family includes executioner caspase-3, which plays a central role in cell apoptosis and differentiation. Unfortunately, there exists a dearth of chemical tools to selectively monitor caspase-3 activity under complex cellular and in vivo conditions due to its close homology with executioner caspase-7. Commercially available activity-based probes and substrates rely on the canonical DEVD tetrapeptide sequence, which both caspases-3 and -7 recognize with similar affinity, and thus the individual contributions of caspase-3 and/or -7 toward important cellular processes are irresolvable. Here, we analyzed a variety of permutations of the DEVD peptide sequence in order to discover peptides with biased activity and recognition of caspase-3 versus caspases-6, -7, -8, and -9. Through this study, we identify fluorescent and biotinylated probes capable of selective detection of caspase-3 using key unnatural amino acids. Likewise, we determined the X-ray crystal structures of caspases-3, -7, and -8 in complex with our lead peptide inhibitor to elucidate the binding mechanism and active site interactions that promote the selective recognition of caspase-3 over other highly homologous caspase family members.
Organizational Affiliation:
Departments of Molecular and Experimental Medicine and Chemical Physiology, The Scripps Research Institute , La Jolla, California 92037, United States.