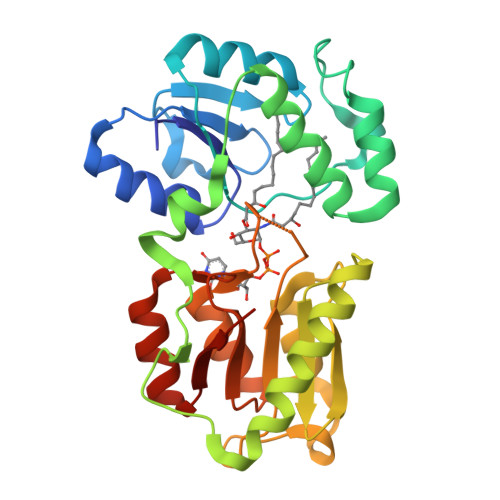
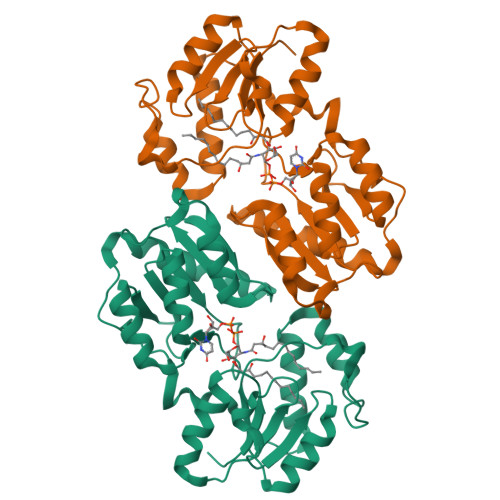
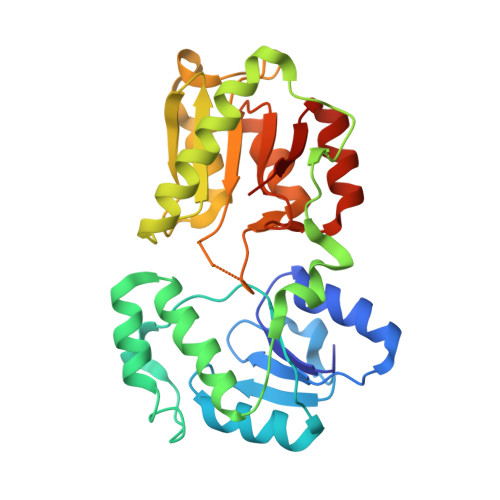
LpxI structures reveal how a lipid A precursor is synthesized.
Metzger IV, L.E., Lee, J.K., Finer-Moore, J.S., Raetz, C.R.H., Stroud, R.M.(2012) Nat Struct Mol Biol 19: 1132-1138
- PubMed: 23042606
- DOI: https://doi.org/10.1038/nsmb.2393
- Primary Citation of Related Structures:
4GGM, 4J6E - PubMed Abstract:
Enzymes in lipid metabolism acquire and deliver hydrophobic substrates and products from within lipid bilayers. The structure at 2.55 Å of one isozyme of a constitutive enzyme in lipid A biosynthesis, LpxI from Caulobacter crescentus, has a novel fold. Two domains close around a completely sequestered substrate, UDP-2,3-diacylglucosamine, and open to release products either to the neighboring enzyme in a putative multienzyme complex or to the bilayer. Mutation analysis identifies Asp225 as key to Mg(2+)-catalyzed diphosphate hydrolysis. These structures provide snapshots of the enzymatic synthesis of a critical lipid A precursor.
Organizational Affiliation:
Department of Biochemistry and Biophysics, The University of California San Francisco, San Francisco, California, USA. metzger@msg.ucsf.edu