Structural and energetic basis of folded-protein transport by the FimD usher.
Geibel, S., Procko, E., Hultgren, S.J., Baker, D., Waksman, G.(2013) Nature 496: 243-246
- PubMed: 23579681
- DOI: https://doi.org/10.1038/nature12007
- Primary Citation of Related Structures:
4J3O - PubMed Abstract:
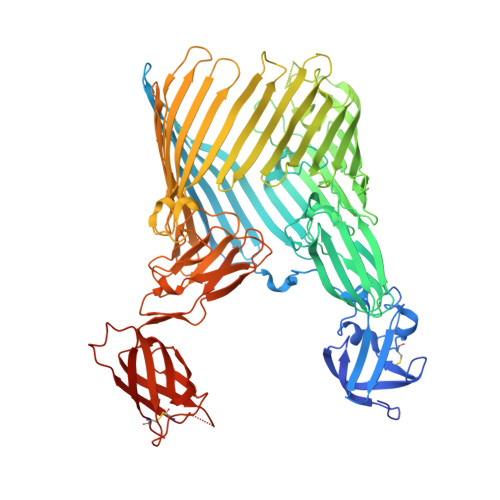
Type 1 pili, produced by uropathogenic Escherichia coli, are multisubunit fibres crucial in recognition of and adhesion to host tissues. During pilus biogenesis, subunits are recruited to an outer membrane assembly platform, the FimD usher, which catalyses their polymerization and mediates pilus secretion. The recent determination of the crystal structure of an initiation complex provided insight into the initiation step of pilus biogenesis resulting in pore activation, but very little is known about the elongation steps that follow. Here, to address this question, we determine the structure of an elongation complex in which the tip complex assembly composed of FimC, FimF, FimG and FimH passes through FimD. This structure demonstrates the conformational changes required to prevent backsliding of the nascent pilus through the FimD pore and also reveals unexpected properties of the usher pore. We show that the circular binding interface between the pore lumen and the folded substrate participates in transport by defining a low-energy pathway along which the nascent pilus polymer is guided during secretion.
Organizational Affiliation:
Institute of Structural and Molecular Biology, University College London and Birkbeck College, Malet Street, London WC1E 7HX, UK.