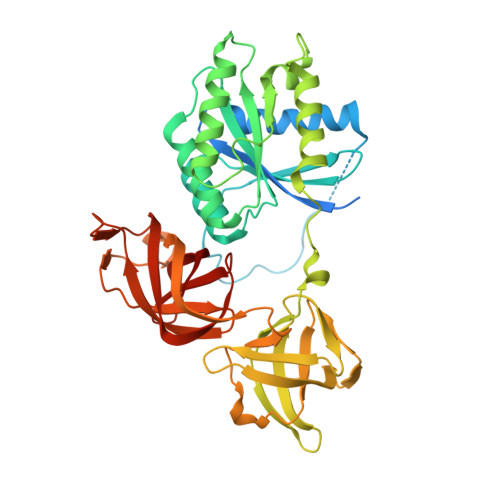
Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation
Scotti, J.S., Leung, I.K.H., Ge, W., Bentley, M.A., Paps, J., Kramer, H.B., Lee, J., Aik, W., Choi, H., Paulsen, S.M., Bowman, L.A.H., Loik, N.D., Horita, S., Ho, C.H., Kershaw, N.J., Tang, C.M., Claridge, T.D.W., Preston, G.M., McDonough, M.A., Schofield, C.J.(2014) Proc Natl Acad Sci U S A 111: 13331-13336
- PubMed: 25197067
- DOI: https://doi.org/10.1073/pnas.1409916111
- Primary Citation of Related Structures:
4IW3, 4J0Q, 4J25 - PubMed Abstract:
The roles of 2-oxoglutarate (2OG)-dependent prolyl-hydroxylases in eukaryotes include collagen stabilization, hypoxia sensing, and translational regulation. The hypoxia-inducible factor (HIF) sensing system is conserved in animals, but not in other organisms. However, bioinformatics imply that 2OG-dependent prolyl-hydroxylases (PHDs) homologous to those acting as sensing components for the HIF system in animals occur in prokaryotes. We report cellular, biochemical, and crystallographic analyses revealing that Pseudomonas prolyl-hydroxylase domain containing protein (PPHD) contain a 2OG oxygenase related in structure and function to the animal PHDs. A Pseudomonas aeruginosa PPHD knockout mutant displays impaired growth in the presence of iron chelators and increased production of the virulence factor pyocyanin. We identify elongation factor Tu (EF-Tu) as a PPHD substrate, which undergoes prolyl-4-hydroxylation on its switch I loop. A crystal structure of PPHD reveals striking similarity to human PHD2 and a Chlamydomonas reinhardtii prolyl-4-hydroxylase. A crystal structure of PPHD complexed with intact EF-Tu reveals that major conformational changes occur in both PPHD and EF-Tu, including a >20-Å movement of the EF-Tu switch I loop. Comparison of the PPHD structures with those of HIF and collagen PHDs reveals conservation in substrate recognition despite diverse biological roles and origins. The observed changes will be useful in designing new types of 2OG oxygenase inhibitors based on various conformational states, rather than active site iron chelators, which make up most reported 2OG oxygenase inhibitors. Structurally informed phylogenetic analyses suggest that the role of prolyl-hydroxylation in human hypoxia sensing has ancient origins.
Organizational Affiliation:
Chemistry Research Laboratory, Department of Chemistry, University of Oxford, Oxford OX1 3TA, United Kingdom;