Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates.
Magee, T.V., Han, S., McCurdy, S.P., Nguyen, T.T., Granskog, K., Marr, E.S., Maguire, B.A., Huband, M.D., Chen, J.M., Subashi, T.A., Shanmugasundaram, V.(2013) Bioorg Med Chem Lett 23: 1727-1731
- PubMed: 23414806
- DOI: https://doi.org/10.1016/j.bmcl.2013.01.067
- Primary Citation of Related Structures:
4IO9, 4IOA, 4IOC - PubMed Abstract:
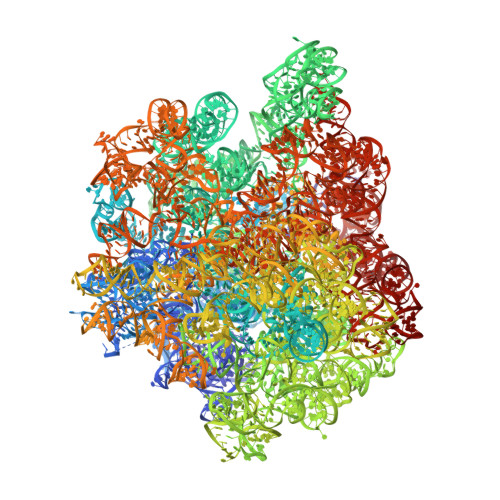
A novel series of 3-O-carbamoyl erythromycin A derived analogs, labeled carbamolides, with activity versus resistant bacterial isolates of staphylococci (including macrolide and oxazolidinone resistant strains) and streptococci are reported. An (R)-2-aryl substituent on a pyrrolidine carbamate appeared to be critical for achieving potency against resistant strains. Crystal structures showed a distinct aromatic interaction between the (R)-2-aryl (3-pyridyl for 4d) substituent on the pyrrolidine and G2484 (G2505, Escherichia coli) of the Deinococcus radiodurans 50S ribosome (3.2Å resolution).
Organizational Affiliation:
Pfizer Cardiovascular, Metabolic, and Endocrine Diseases (CVMED) Chemistry, 620 Memorial Dr, Cambridge, MA 02139, USA. thomas.v.magee@pfizer.com