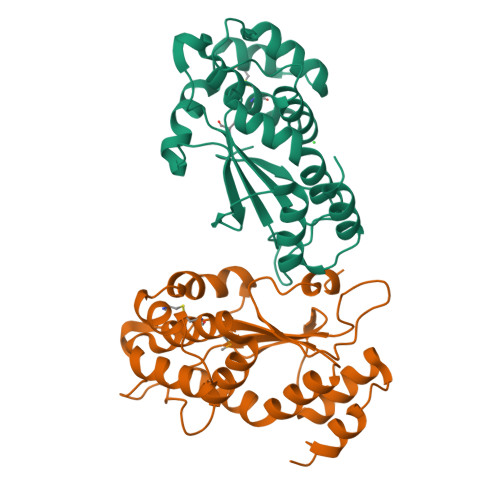
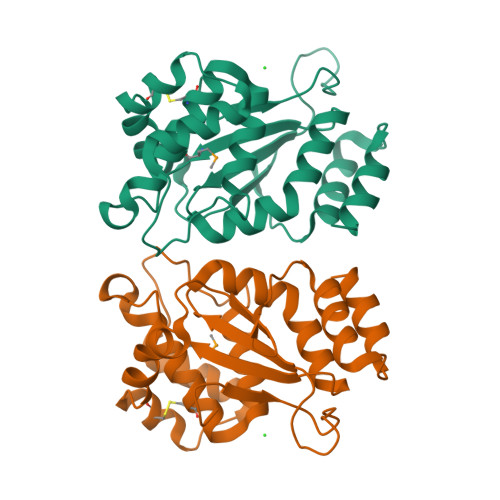
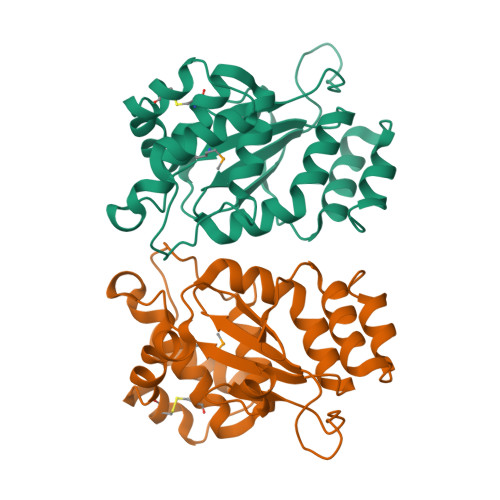
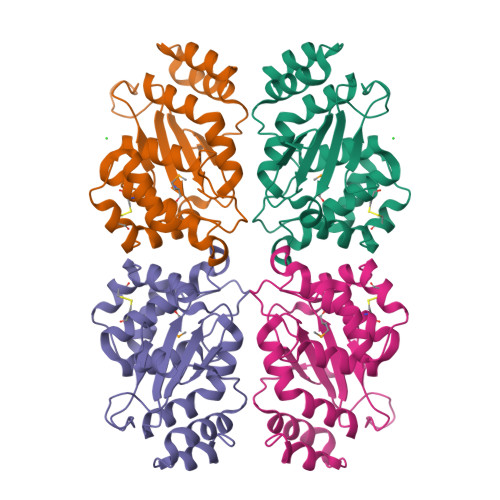
Structural and biochemical characterization of the essential DsbA-like disulfide bond forming protein from Mycobacterium tuberculosis.
Chim, N., Harmston, C.A., Guzman, D.J., Goulding, C.W.(2013) BMC Struct Biol 13: 23-23
- PubMed: 24134223
- DOI: https://doi.org/10.1186/1472-6807-13-23
- Primary Citation of Related Structures:
4IHU - PubMed Abstract:
Bacterial Disulfide bond forming (Dsb) proteins facilitate proper folding and disulfide bond formation of periplasmic and secreted proteins. Previously, we have shown that Mycobacterium tuberculosis Mt-DsbE and Mt-DsbF aid in vitro oxidative folding of proteins. The M. tuberculosis proteome contains another predicted membrane-tethered Dsb protein, Mt-DsbA, which is encoded by an essential gene. Herein, we present structural and biochemical analyses of Mt-DsbA. The X-ray crystal structure of Mt-DsbA reveals a two-domain structure, comprising a canonical thioredoxin domain with the conserved CXXC active site cysteines in their reduced form, and an inserted α-helical domain containing a structural disulfide bond. The overall fold of Mt-DsbA resembles that of other DsbA-like proteins and not Mt-DsbE or Mt-DsbF. Biochemical characterization demonstrates that, unlike Mt-DsbE and Mt-DsbF, Mt-DsbA is unable to oxidatively fold reduced, denatured hirudin. Moreover, on the substrates tested in this study, Mt-DsbA has disulfide bond isomerase activity contrary to Mt-DsbE and Mt-DsbF. These results suggest that Mt-DsbA acts upon a distinct subset of substrates as compared to Mt-DsbE and Mt-DsbF. One could speculate that Mt-DsbE and Mt-DsbF are functionally redundant whereas Mt-DsbA is not, offering an explanation for the essentiality of Mt-DsbA in M. tuberculosis.
Organizational Affiliation:
Departments of Molecular Biology and Biochemistry, UCI, Irvine, CA 92697, USA. celia.goulding@uci.edu.