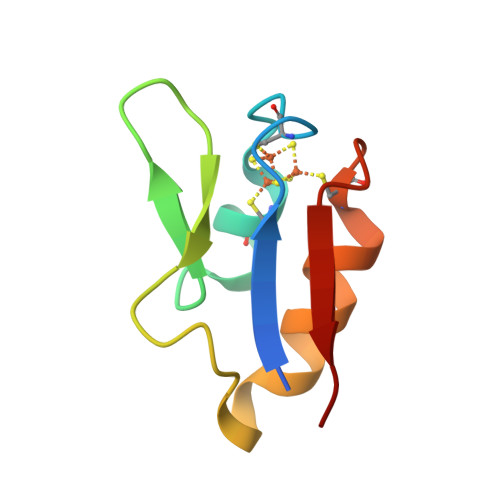
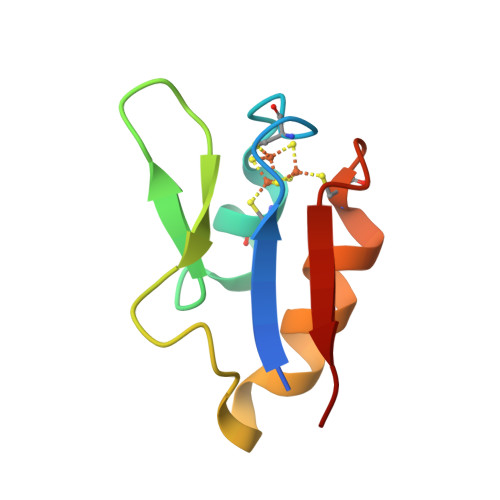
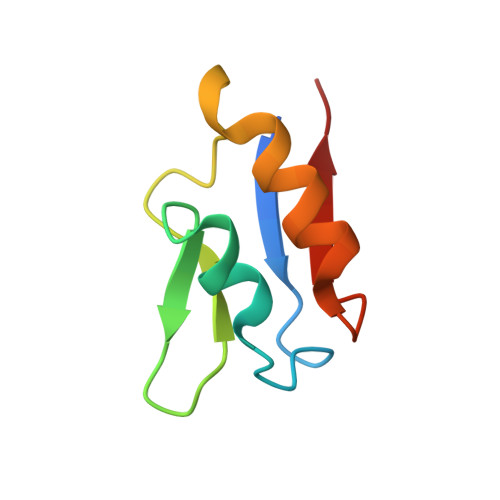
The structure of a novel electron-transfer ferredoxin from Rhodopseudomonas palustris HaA2 which contains a histidine residue in its iron-sulfur cluster-binding motif.
Zhang, T., Zhang, A., Bell, S.G., Wong, L.L., Zhou, W.(2014) Acta Crystallogr D Biol Crystallogr 70: 1453-1464
- PubMed: 24816113
- DOI: https://doi.org/10.1107/S139900471400474X
- Primary Citation of Related Structures:
4ID8, 4OV1 - PubMed Abstract:
Rhodopseudomonas palustris HaA2 contains a gene, RPB3630, encoding a ferredoxin, HaPuxC, with an atypical CXXHXXC(X)nCP iron-sulfur cluster-binding motif. The ferredoxin gene is associated with a cytochrome P450 (CYP) monooxygenase-encoding gene, CYP194A3, an arrangement which is conserved in several strains of bacteria. Similar ferredoxin genes are found in other bacteria, such as Mycobacterium tuberculosis, where they are also associated with CYP genes. The crystal structure of HaPuxC has been solved at 2.3 Å resolution. The overall fold of this [3Fe-4S] cluster-containing ferredoxin is similar to other [3Fe-4S] and [4Fe-4S] species, with the loop around the iron-sulfur cluster more closely resembling those of [3Fe-4S] ferredoxins. The side chain of His17 from the cluster-binding motif in HaPuxC points away from the vacant site of the cluster and interacts with Glu61 and one of the sulfide ions of the cluster. This is the first cytochrome P450 electron-transfer partner of this type to be structurally characterized and will provide a better understanding of the electron-transfer processes between these ferredoxins and their CYP enzymes.
Organizational Affiliation:
College of Life Sciences, Nankai University, Tianjin 300071, People's Republic of China.