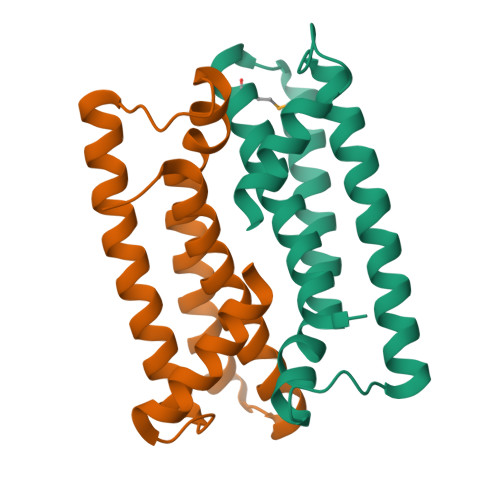
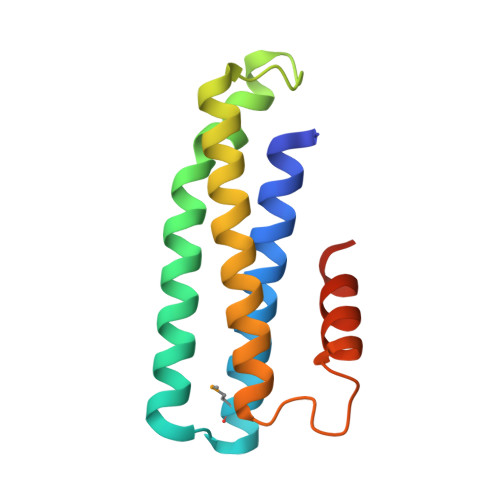
Crystal structure of PAV1-137: a protein from the virus PAV1 that infects Pyrococcus abyssi
Leulliot, N., Quevillon-Cheruel, S., Graille, M., Geslin, C., Flament, D., Romancer, M.L., Tilbeurgh, H.V.(2013) Archaea 2013: 568053-568053
- PubMed: 23533329
- DOI: https://doi.org/10.1155/2013/568053
- Primary Citation of Related Structures:
4HR1 - PubMed Abstract:
Pyrococcus abyssi virus 1 (PAV1) was the first virus particle infecting a hyperthermophilic Euryarchaeota (Pyrococcus abyssi strain GE23) that has been isolated and characterized. It is lemon shaped and is decorated with a short fibered tail. PAV1 morphologically resembles the fusiform members of the family Fuselloviridae or the genus Salterprovirus. The 18 kb dsDNA genome of PAV1 contains 25 predicted genes, most of them of unknown function. To help assigning functions to these proteins, we have initiated structural studies of the PAV1 proteome. We determined the crystal structure of a putative protein of 137 residues (PAV1-137) at a resolution of 2.2 Å. The protein forms dimers both in solution and in the crystal. The fold of PAV1-137 is a four- α -helical bundle analogous to those found in some eukaryotic adhesion proteins such as focal adhesion kinase, suggesting that PAV1-137 is involved in protein-protein interactions.
Organizational Affiliation:
Institut de Biochimie et de Biophysique Moléculaire et Cellulaire, CNRS-UMR 8619, IFR115, Université Paris-Sud, Bâtiment 430, 91405 Orsay, France.