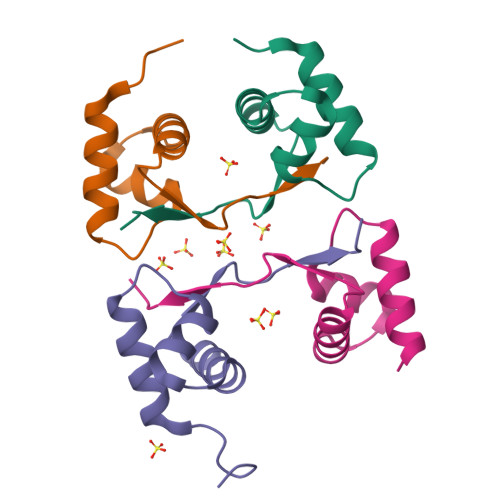
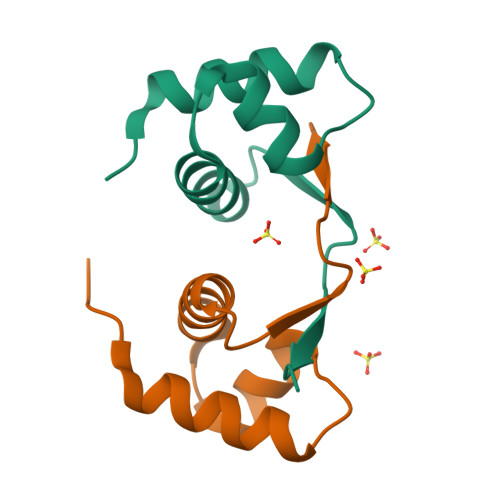
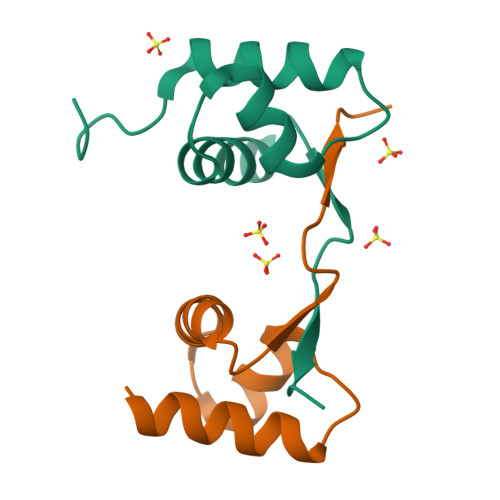
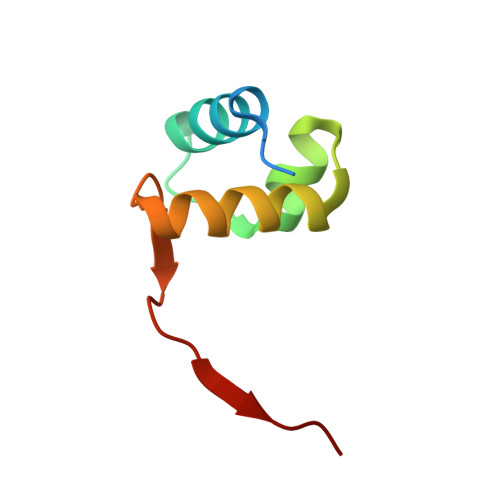
Crystal structure of a poxvirus-like zalpha domain from cyprinid herpesvirus 3
Tome, A.R., Kus, K., Correia, S., Paulo, L.M., Zacarias, S., de Rosa, M., Figueiredo, D., Parkhouse, R.M., Athanasiadis, A.(2013) J Virol 87: 3998-4004
- PubMed: 23365431
- DOI: https://doi.org/10.1128/JVI.03116-12
- Primary Citation of Related Structures:
4HOB - PubMed Abstract:
Zalpha domains are a subfamily of the winged helix-turn-helix domains sharing the unique ability to recognize CpG repeats in the left-handed Z-DNA conformation. In vertebrates, domains of this family are found exclusively in proteins that detect foreign nucleic acids and activate components of the antiviral interferon response. Moreover, poxviruses encode the Zalpha domain-containing protein E3L, a well-studied and potent inhibitor of interferon response. Here we describe a herpesvirus Zalpha-domain-containing protein (ORF112) from cyprinid herpesvirus 3. We demonstrate that ORF112 also binds CpG repeats in the left-handed conformation, and moreover, its structure at 1.75 Å reveals the Zalpha fold found in ADAR1, DAI, PKZ, and E3L. Unlike other Zalpha domains, however, ORF112 forms a dimer through a unique domain-swapping mechanism. Thus, ORF112 may be considered a new member of the Z-domain family having DNA binding properties similar to those of the poxvirus E3L inhibitor of interferon response.
Organizational Affiliation:
Instituto Gulbenkian de Ciência, Oeiras, Portugal.