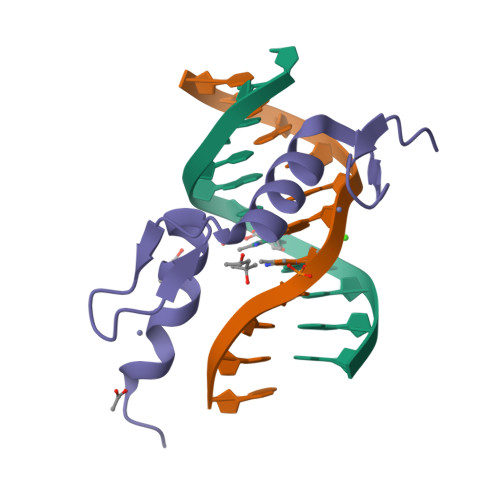
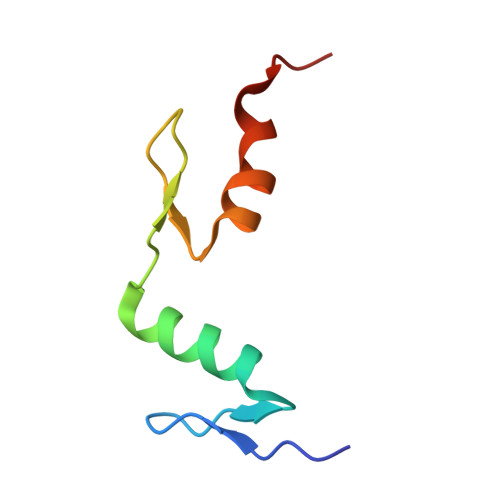
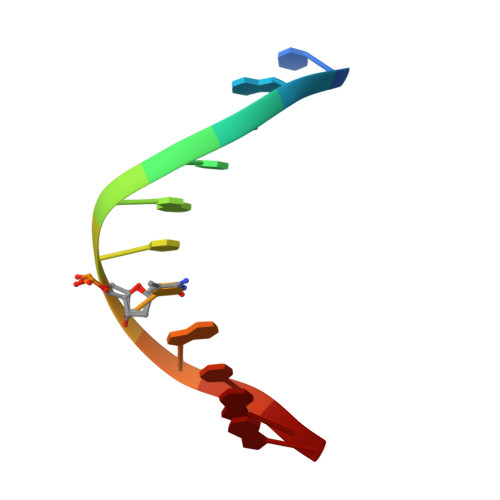
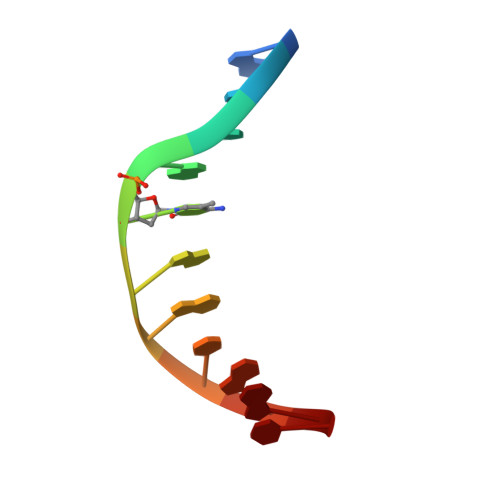
An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence.
Liu, Y., Toh, H., Sasaki, H., Zhang, X., Cheng, X.(2012) Genes Dev 26: 2374-2379
- PubMed: 23059534
- DOI: https://doi.org/10.1101/gad.202200.112
- Primary Citation of Related Structures:
4GZN - PubMed Abstract:
Zinc finger transcription factor Zfp57 recognizes the methylated CpG within the TGCCGC element. We determined the structure of the DNA-binding domain of Zfp57, consisting of two adjacent zinc fingers, in complex with fully methylated DNA at 1.0 Å resolution. The first zinc finger contacts the 5' half (TGC), and the second recognizes the 3' half (CGC) of the recognition sequence. Zfp57 recognizes the two 5-methylcytosines (5mCs) asymmetrically: One involves hydrophobic interactions with Arg178, which also interacts with the neighboring 3' guanine and forms a 5mC-Arg-G interaction, while the other involves a layer of ordered water molecules. Two point mutations in patients with transient neonatal diabetes abolish DNA-binding activity. Zfp57 has reduced binding affinity for unmodified DNA and the oxidative products of 5mC.
Organizational Affiliation:
Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.