Structure and function of non-native metal clusters in human arginase I.
D'Antonio, E.L., Hai, Y., Christianson, D.W.(2012) Biochemistry 51: 8399-8409
- PubMed: 23061982
- DOI: https://doi.org/10.1021/bi301145n
- Primary Citation of Related Structures:
4GSM, 4GSV, 4GSZ, 4GWC, 4GWD - PubMed Abstract:
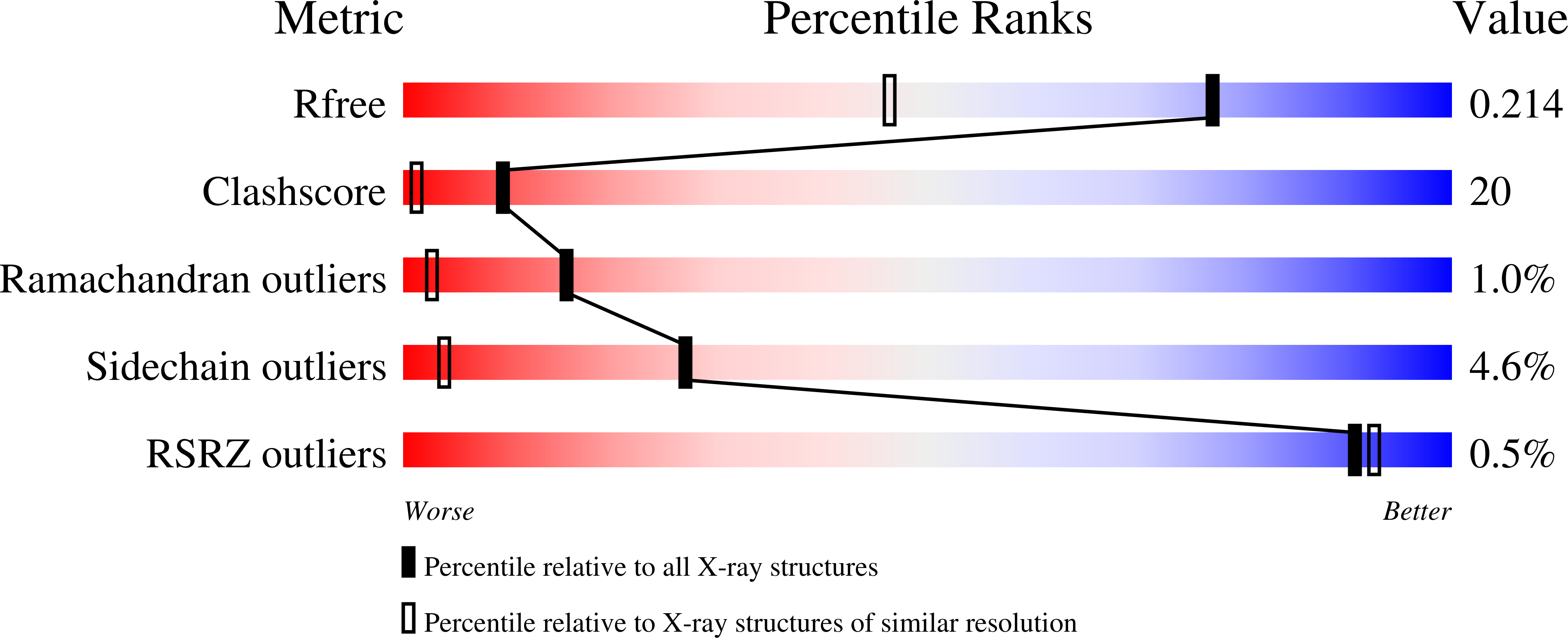
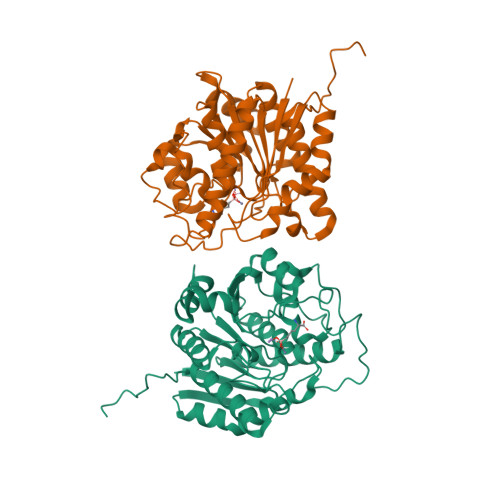
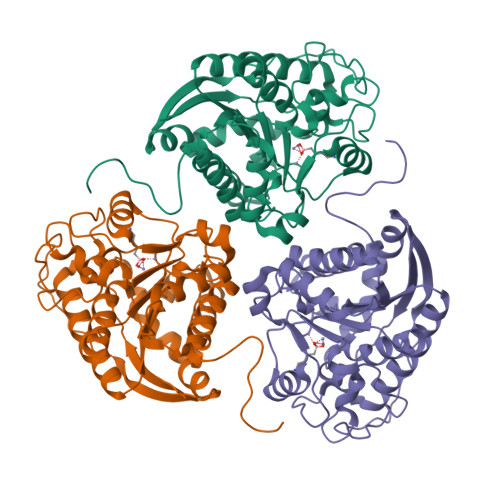
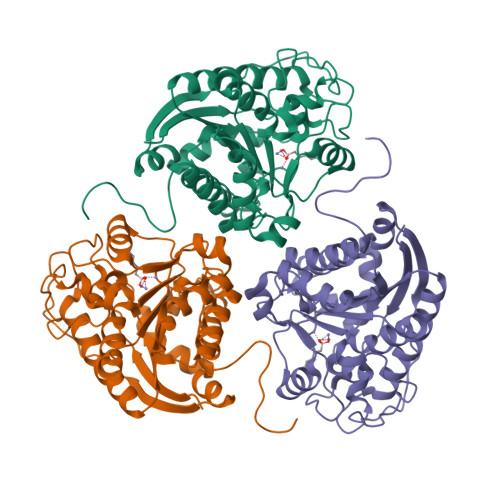
Various binuclear metal ion clusters and complexes have been reconstituted in crystalline human arginase I by removing the Mn(2+)(2) cluster of the wild-type enzyme with metal chelators and subsequently soaking the crystalline apoenzyme in buffer solutions containing NiCl(2) or ZnCl(2). X-ray crystal structures of these metal ion variants are correlated with catalytic activity measurements that reveal differences resulting from metal ion substitution. Additionally, treatment of crystalline Mn(2+)(2)-human arginase I with Zn(2+) reveals for the first time the structural basis for inhibition by Zn(2+), which forms a carboxylate-histidine-Zn(2+) triad with H141 and E277. The imidazole side chain of H141 is known to be hyper-reactive, and its chemical modification or mutagenesis is known to similarly compromise catalysis. The reactive substrate analogue 2(S)-amino-6-boronohexanoic acid (ABH) binds as a tetrahedral boronate anion to Mn(2+)(2), Co(2+)(2), Ni(2+)(2), and Zn(2+)(2) clusters in human arginase I, and it can be stabilized by a third inhibitory Zn(2+) ion coordinated by H141. Because ABH binds as an analogue of the tetrahedral intermediate and its flanking transition states in catalysis, this implies that the various metallo-substituted enzymes are capable of some level of catalysis with an actual substrate. Accordingly, we establish the following trend for turnover number (k(cat)) and catalytic efficiency (k(cat)/K(M)): Mn(2+) > Ni(2+) ≈ Co(2+) ≫ Zn(2+). Therefore, Mn(2+) is required for optimal catalysis by human arginase I.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104-6323, USA.