Crystal structure of the omega-aminotransferase from Paracoccus denitrificans and its phylogenetic relationship with other class III aminotransferases that have biotechnological potential.
Rausch, C., Lerchner, A., Schiefner, A., Skerra, A.(2013) Proteins 81: 774-787
- PubMed: 23239223
- DOI: https://doi.org/10.1002/prot.24233
- Primary Citation of Related Structures:
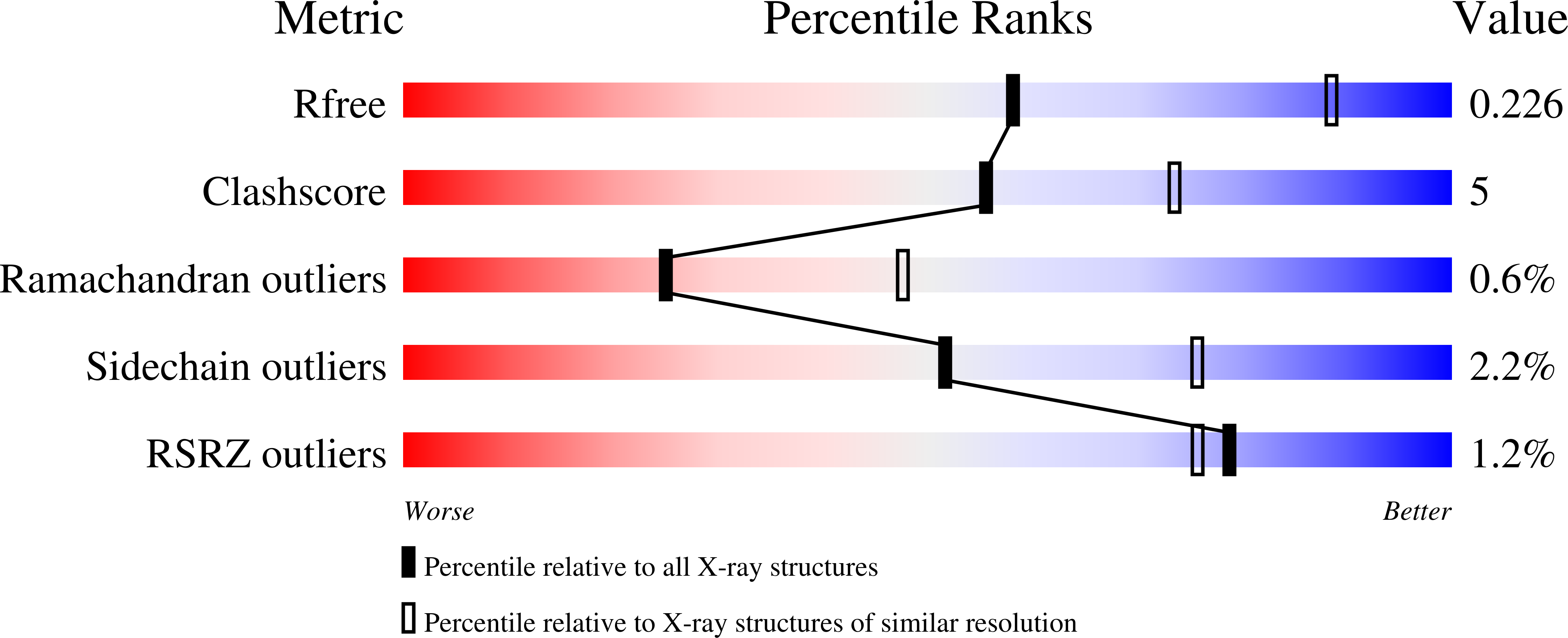
4GRX - PubMed Abstract:
Apart from their crucial role in metabolism, pyridoxal 5'-phosphate (PLP)-dependent aminotransferases (ATs) constitute a class of enzymes with increasing application in industrial biotechnology. To provide better insight into the structure-function relationships of ATs with biotechnological potential we performed a fundamental bioinformatics analysis of 330 representative sequences of pro- and eukaryotic Class III ATs using a structure-guided approach. The calculated phylogenetic maximum likelihood tree revealed six distinct clades of which the first segregates with a very high bootstrap value of 92%. Most enzymes in this first clade have been functionally well characterized, whereas knowledge about the natural functions and substrates of enzymes in the other branches is sparse. Notably, in those clades 2-6 members of the peculiar class of ω-ATs prevail, many of which have proven useful for the preparation of chiral amines or artificial amino acids. One representative is the ω-AT from Paracoccus denitrificans (PD ω-AT) which catalyzes, for example, the transamination in a novel biocatalytic process for the production of L-homoalanine from L-threonine. To gain structural insight into this important enzyme, its X-ray analysis was carried out at a resolution of 2.6 Å, including the covalently bound PLP as well as 5-aminopentanoate as a putative amino donor substrate. On the basis of this crystal structure in conjunction with our phylogenetic analysis, we have identified a generic set of active site residues of ω-ATs that are associated with a strong preference for aromatic substrates, thus guiding the discovery of novel promising enzymes for the biotechnological production of corresponding chiral amines.
Organizational Affiliation:
Munich Center for integrated Protein Science (CiPSM) and Lehrstuhl für Biologische Chemie, Technische Universität München, Freising-Weihenstephan, Germany.