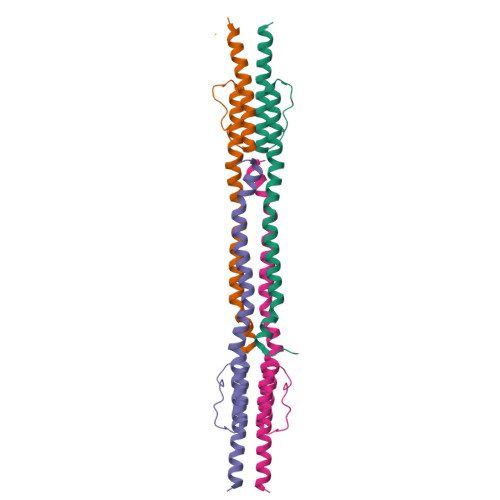
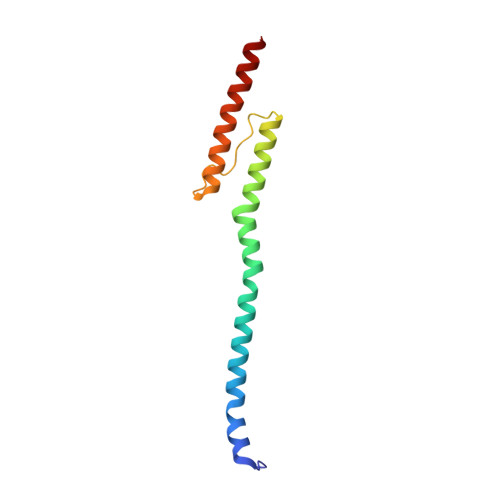
Exploiting tertiary structure through local folds for crystallographic phasing.
Sammito, M., Millan, C., Rodriguez, D.D., de Ilarduya, I.M., Meindl, K., De Marino, I., Petrillo, G., Buey, R.M., de Pereda, J.M., Zeth, K., Sheldrick, G.M., Uson, I.(2013) Nat Methods 10: 1099-1101
- PubMed: 24037245
- DOI: https://doi.org/10.1038/nmeth.2644
- Primary Citation of Related Structures:
4GDO, 4GN0 - PubMed Abstract:
We describe an algorithm for phasing protein crystal X-ray diffraction data that identifies, retrieves, refines and exploits general tertiary structural information from small fragments available in the Protein Data Bank. The algorithm successfully phased, through unspecific molecular replacement combined with density modification, all-helical, mixed alpha-beta, and all-beta protein structures. The method is available as a software implementation: Borges.
Organizational Affiliation:
Instituto de Biología Molecular de Barcelona, Consejo Superior de Investigaciones Científicas, Barcelona, Spain.