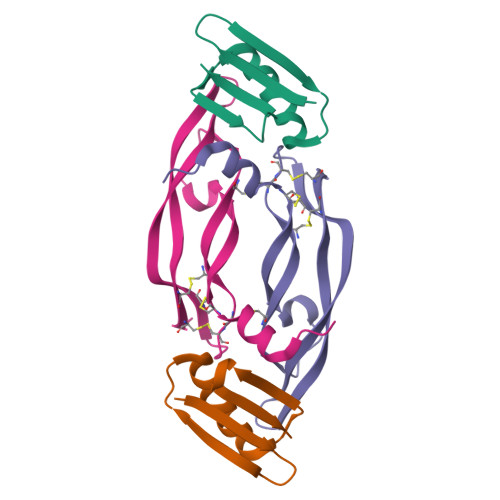
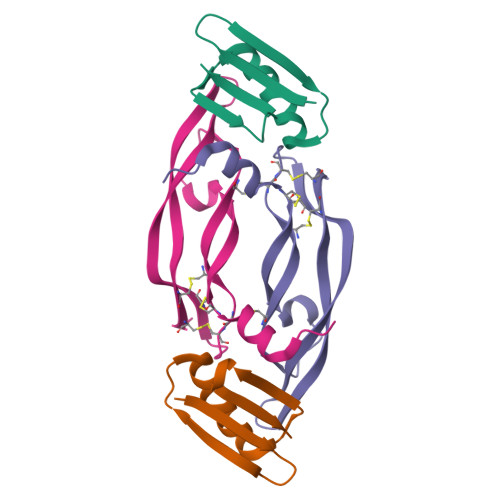
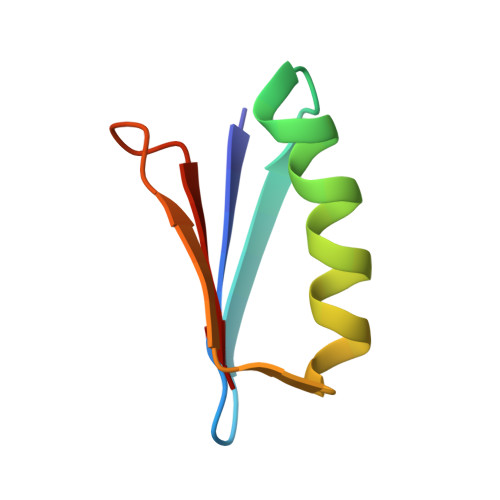
Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Mandal, K., Uppalapati, M., Ault-Riche, D., Kenney, J., Lowitz, J., Sidhu, S.S., Kent, S.B.(2012) Proc Natl Acad Sci U S A 109: 14779-14784
- PubMed: 22927390
- DOI: https://doi.org/10.1073/pnas.1210483109
- Primary Citation of Related Structures:
4GLN, 4GLS, 4GLU - PubMed Abstract:
Total chemical synthesis was used to prepare the mirror image (D-protein) form of the angiogenic protein vascular endothelial growth factor (VEGF-A). Phage display against D-VEGF-A was used to screen designed libraries based on a unique small protein scaffold in order to identify a high affinity ligand. Chemically synthesized D- and L- forms of the protein ligand showed reciprocal chiral specificity in surface plasmon resonance binding experiments: The L-protein ligand bound only to D-VEGF-A, whereas the D-protein ligand bound only to L-VEGF-A. The D-protein ligand, but not the L-protein ligand, inhibited the binding of natural VEGF(165) to the VEGFR1 receptor. Racemic protein crystallography was used to determine the high resolution X-ray structure of the heterochiral complex consisting of {D-protein antagonist + L-protein form of VEGF-A}. Crystallization of a racemic mixture of these synthetic proteins in appropriate stoichiometry gave a racemic protein complex of more than 73 kDa containing six synthetic protein molecules. The structure of the complex was determined to a resolution of 1.6 Å. Detailed analysis of the interaction between the D-protein antagonist and the VEGF-A protein molecule showed that the binding interface comprised a contact surface area of approximately 800 Å(2) in accord with our design objectives, and that the D-protein antagonist binds to the same region of VEGF-A that interacts with VEGFR1-domain 2.
Organizational Affiliation:
Department of Chemistry, Institute for Biophysical Dynamics, University of Chicago, Chicago, IL 60637, USA.