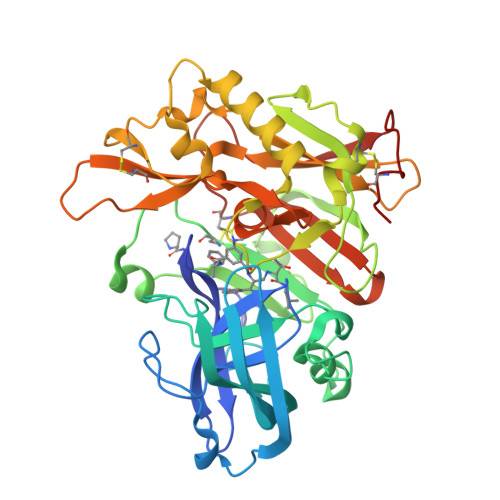
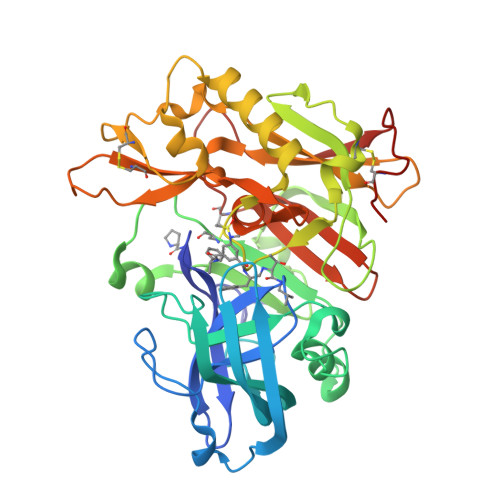
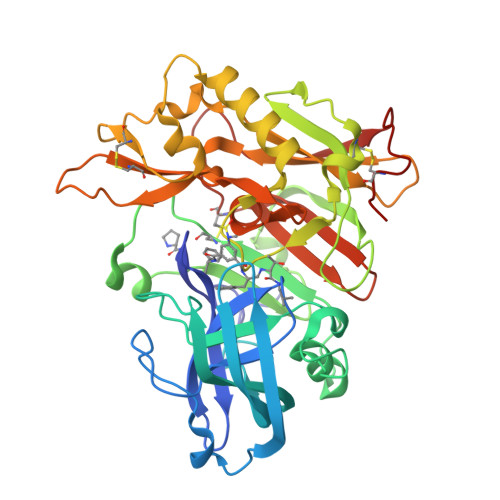
Structure-based design of highly selective beta-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure.
Ghosh, A.K., Venkateswara Rao, K., Yadav, N.D., Anderson, D.D., Gavande, N., Huang, X., Terzyan, S., Tang, J.(2012) J Med Chem 55: 9195-9207
- PubMed: 22954357
- DOI: https://doi.org/10.1021/jm3008823
- Primary Citation of Related Structures:
4GID - PubMed Abstract:
The structure-based design, synthesis, and X-ray structure of protein-ligand complexes of exceptionally potent and selective β-secretase inhibitors are described. The inhibitors are designed specifically to interact with S(1)' active site residues to provide selectivity over memapsin 1 and cathepsin D. Inhibitor 5 has exhibited exceedingly potent inhibitory activity (K(i) = 17 pM) and high selectivity over BACE 2 (>7000-fold) and cathepsin D (>250000-fold). A protein-ligand crystal structure revealed important molecular insight into these selectivities. These interactions may serve as an important guide to design selectivity over the physiologically important aspartic acid proteases.
Organizational Affiliation:
Department of Chemistry, Purdue University, 560 Oval Drive, West Lafayette, Indiana 47907, United States. akghosh@purdue.edu