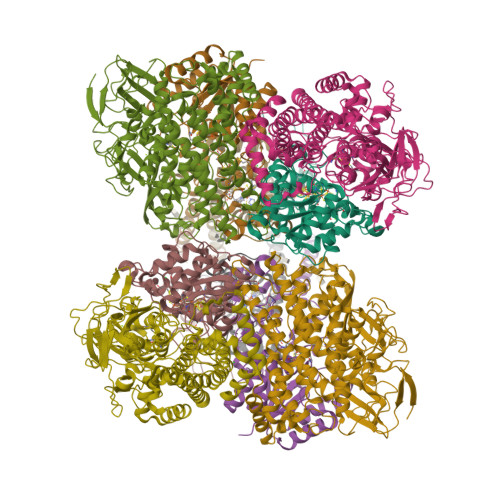
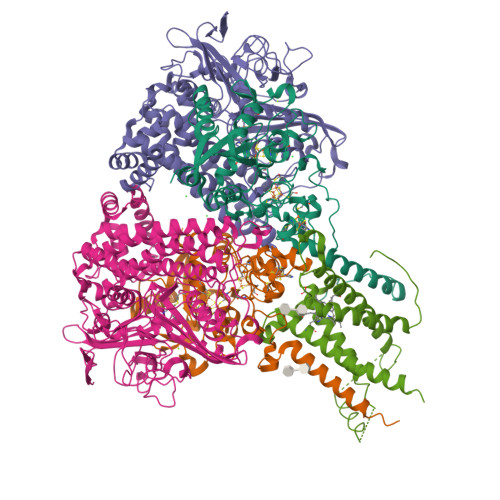
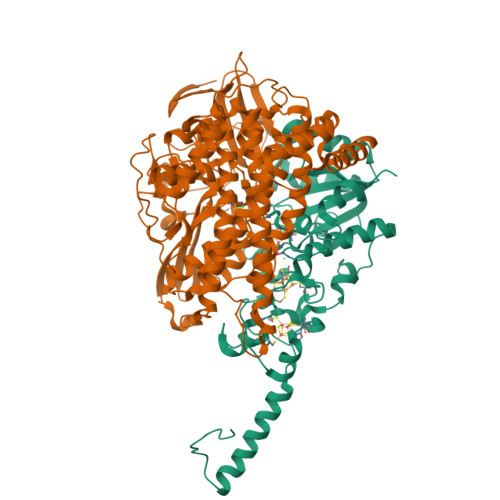
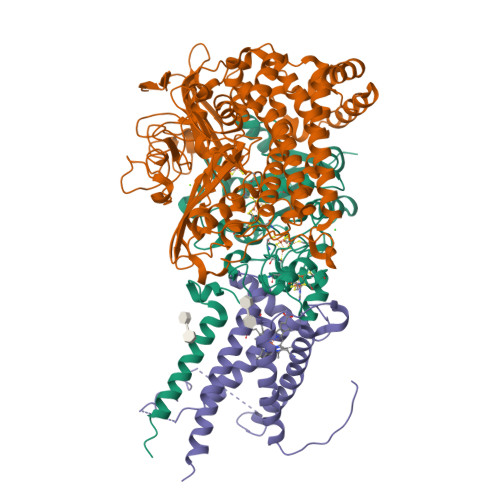
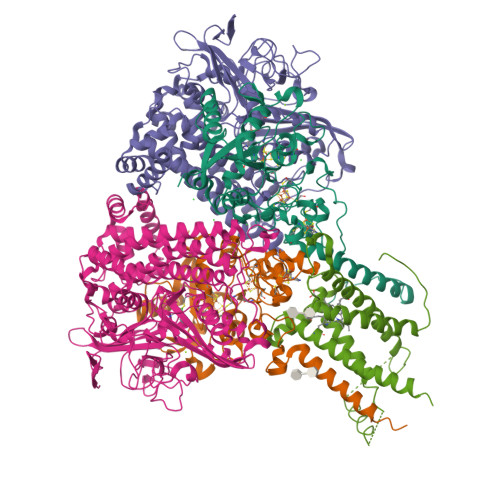
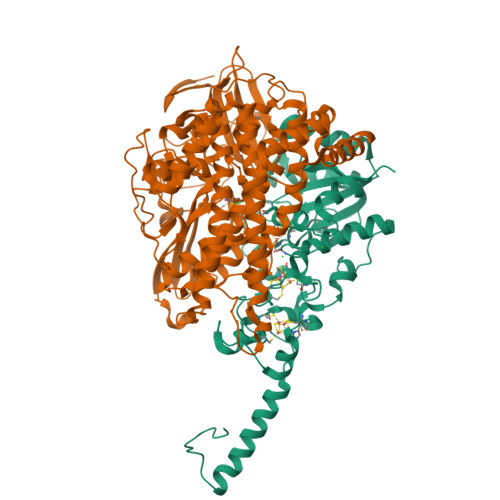
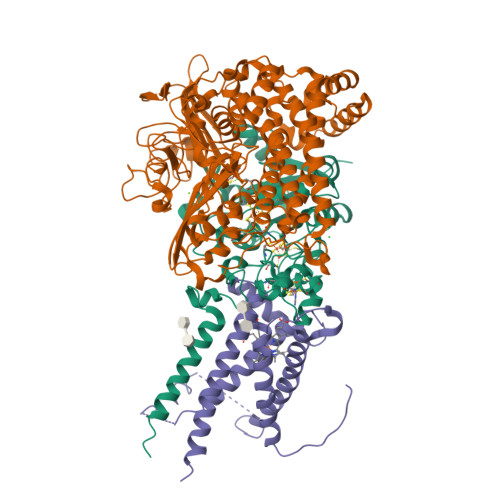
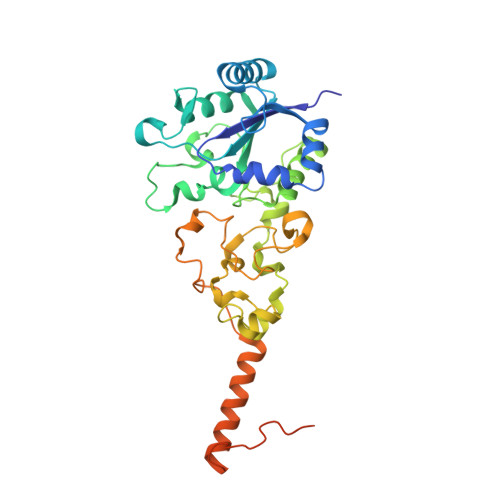
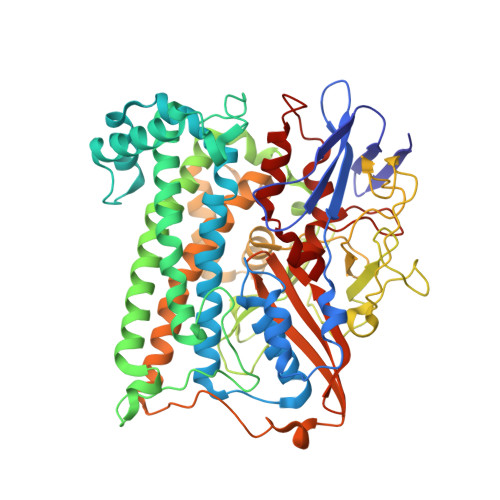
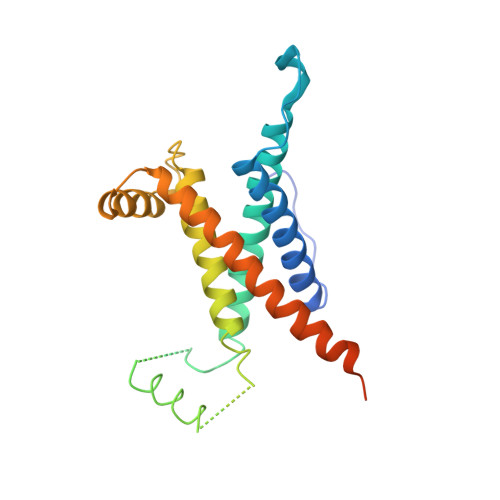
Crystal Structure of the O(2)-Tolerant Membrane-Bound Hydrogenase 1 from Escherichia coli in Complex with Its Cognate Cytochrome b.
Volbeda, A., Darnault, C., Parkin, A., Sargent, F., Armstrong, F.A., Fontecilla-Camps, J.C.(2013) Structure 21: 184-190
- PubMed: 23260654
- DOI: https://doi.org/10.1016/j.str.2012.11.010
- Primary Citation of Related Structures:
4GD3 - PubMed Abstract:
We report the 3.3 Å resolution structure of dimeric membrane-bound O(2)-tolerant hydrogenase 1 from Escherichia coli in a 2:1 complex with its physiological partner, cytochrome b. From the short distance between distal [Fe(4)S(4)] clusters, we predict rapid transfer of H(2)-derived electrons between hydrogenase heterodimers. Thus, under low O(2) levels, a functional active site in one heterodimer can reductively reactivate its O(2)-exposed counterpart in the other. Hydrogenase 1 is maximally expressed during fermentation, when electron acceptors are scarce. These conditions are achieved in the lower part of the host's intestinal tract when E. coli is soon to be excreted and undergo an anaerobic-to-aerobic metabolic transition. The apparent paradox of having an O(2)-tolerant hydrogenase expressed under anoxia makes sense if the enzyme functions to keep intracellular O(2) levels low by reducing it to water, protecting O(2)-sensitive enzymes during the transition. Cytochrome b's main role may be anchoring the hydrogenase to the membrane.
Organizational Affiliation:
Metalloproteins Unit, Institut de Biologie Structurale J.-P. Ebel CEA-CNRS-Université Joseph Fourier, UMR 5075, 41 rue Jules Horowitz, 38027 Grenoble, France.