Antimalarial and Structural Studies of Pyridine-containing Inhibitors of 1-Deoxyxylulose-5-phosphate Reductoisomerase.
Xue, J., Diao, J., Cai, G., Deng, L., Zheng, B., Yao, Y., Song, Y.(2013) ACS Med Chem Lett 4: 278-282
- PubMed: 23795240
- DOI: https://doi.org/10.1021/ml300419r
- Primary Citation of Related Structures:
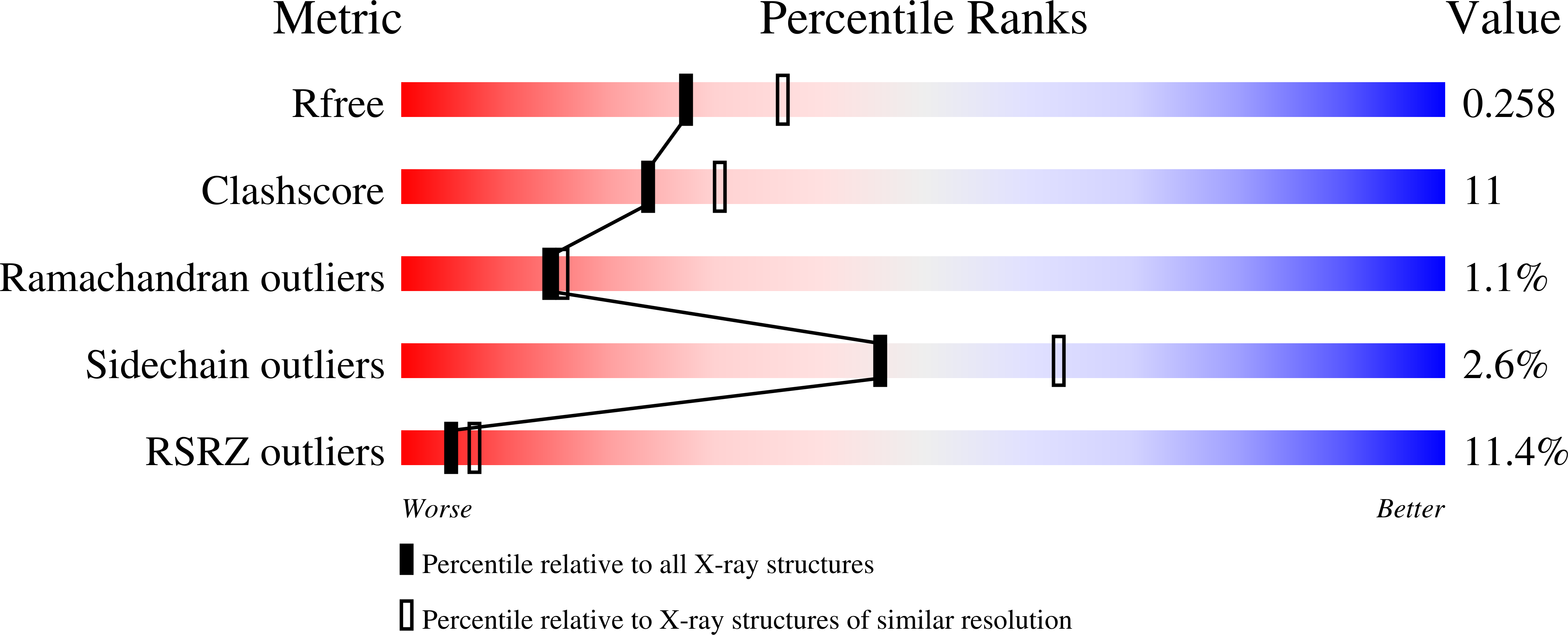
4GAE - PubMed Abstract:
1-Deoxy- D -xylulose-5-phosphate reductoisomerase (DXR) in the non-mevalonate isoprene biosynthesis pathway is a target for developing antimalarial drugs. Fosmidomycin, a potent DXR inhibitor, showed safety as well as efficacy against P. falciparum malaria in clinical trials. Based on our previous quantitative structure activity relationship (QSAR) and crystallographic studies, several novel pyridine-containing fosmidomycin derivatives were designed, synthesized and found to be highly potent inhibitors of P. falciparum DXR ( Pf DXR) having K i values of 1.9 - 13 nM, with the best one being ~11× more active than fosmidomycin. These compounds also potently block the proliferation of multi-drug resistant P. falciparum with EC 50 values as low as 170 nM. A 2.3 Å crystal structure of Pf DXR in complex with one of the inhibitors is reported, showing the flexible loop of the protein undergoes conformational changes upon ligand binding and a hydrogen bond and favorable hydrophobic interactions between the pyridine group and Pf DXR account for the enhanced activity.
Organizational Affiliation:
Department of Pharmacology, Baylor College of Medicine, 1 Baylor Plaza, Houston, Texas 77030, United States.