Product formation controlled by substrate dynamics in leukotriene A4 hydrolase.
Stsiapanava, A., Tholander, F., Kumar, R.B., Qureshi, A.A., Niegowski, D., Hasan, M., Thunnissen, M., Haeggstrom, J.Z., Rinaldo-Matthis, A.(2014) Biochim Biophys Acta 1844: 439-446
- PubMed: 24333438
- DOI: https://doi.org/10.1016/j.bbapap.2013.12.003
- Primary Citation of Related Structures:
4GAA - PubMed Abstract:
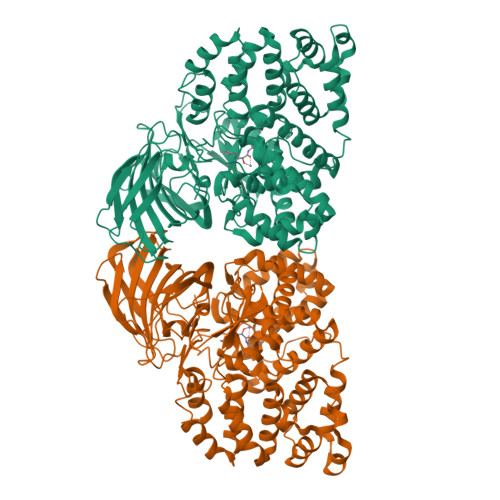
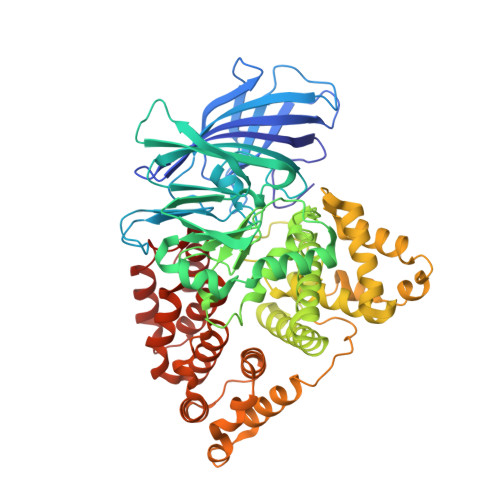
Leukotriene A4 hydrolase/aminopeptidase (LTA4H) (EC 3.3.2.6) is a bifunctional zinc metalloenzyme with both an epoxide hydrolase and an aminopeptidase activity. LTA4H from the African claw toad, Xenopus laevis (xlLTA4H) has been shown to, unlike the human enzyme, convert LTA4 to two enzymatic metabolites, LTB4 and another biologically active product Δ(6)-trans-Δ(8)-cis-LTB4 (5(S),12R-dihydroxy-6,10-trans-8,14-cis-eicosatetraenoic acid). In order to study the molecular aspect of the formation of this product we have characterized the structure and function of xlLTA4H. We solved the structure of xlLTA4H to a resolution of 2.3Å. It is a dimeric structure where each monomer has three domains with the active site in between the domains, similar as to the human structure. An important difference between the human and amphibian enzyme is the phenylalanine to tyrosine exchange at position 375. Our studies show that mutating F375 in xlLTA4H to tyrosine abolishes the formation of the LTB4 isomeric product Δ(6)-trans-Δ(8)-cis-LTB4. In an attempt to understand how one amino acid exchange leads to a new product profile as seen in the xlLTA4H, we performed a conformer analysis of the triene part of the substrate LTA4. Our results show that the Boltzmann distribution of substrate conformers correlates with the observed distribution of products. We suggest that the observed difference in product profile between the human and the xlLTA4H arises from different level of discrimination between substrate LTA4 conformers.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Scheeles väg 2, Karolinska Institutet, 17177 Stockholm, Sweden. Electronic address: alena.stsiapanava@ki.se.