Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Madrona, Y., Tripathi, S., Li, H., Poulos, T.L.(2012) Biochemistry 51: 6623-6631
- PubMed: 22775403
- DOI: https://doi.org/10.1021/bi300666u
- Primary Citation of Related Structures:
4FB2, 4FMX, 4FYZ, 4G3R - PubMed Abstract:
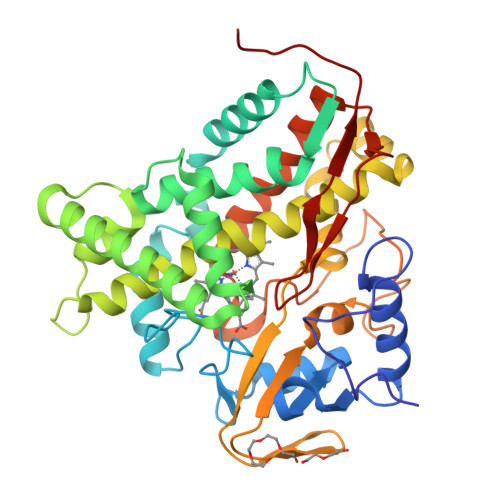
The crystal structure of the P450cin substrate-bound nitric oxide complex and the substrate-free form have been determined revealing a substrate-free structure that adopts an open conformation relative to the substrate-bound structure. The region of the I helix that forms part of the O(2) binding pocket shifts from an α helix in the substrate-free form to a π helix in the substrate-bound form. Unique to P450cin is an active site residue, Asn242, in the I helix that H-bonds with the substrate. In most other P450s this residue is a Thr and plays an important role in O(2) activation by participating in an H-bonding network required for O(2) activation. The π/α I helix transition results in the carbonyl O atom of Gly238 moving in to form an H-bond with the water/hydroxide ligand in the substrate-free form. The corresponding residue, Gly248, in the substrate-free P450cam structure experiences a similar motion. Most significantly, in the oxy-P450cam complex Gly248 adopts a position midway between the substrate-free and -bound states. A comparison between these P450cam and the new P450cin structures provides insights into differences in how the two P450s activate O(2). The structure of P450cin complexed with nitric oxide, a close mimic of the O(2) complex, shows that Gly238 is likely to form tighter interactions with ligands than the corresponding Gly248 in P450cam. Having a close interaction between an H-bond acceptor, the Gly238 carbonyl O atom, and the distal oxygen atom of O(2) will promote protonation and hence further reduction of the oxy complex to the hydroperoxy intermediate resulting in heterolytic cleavage of the peroxide O-O bond and formation of the active ferryl intermediate required for substrate hydroxylation.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697-3900, USA.