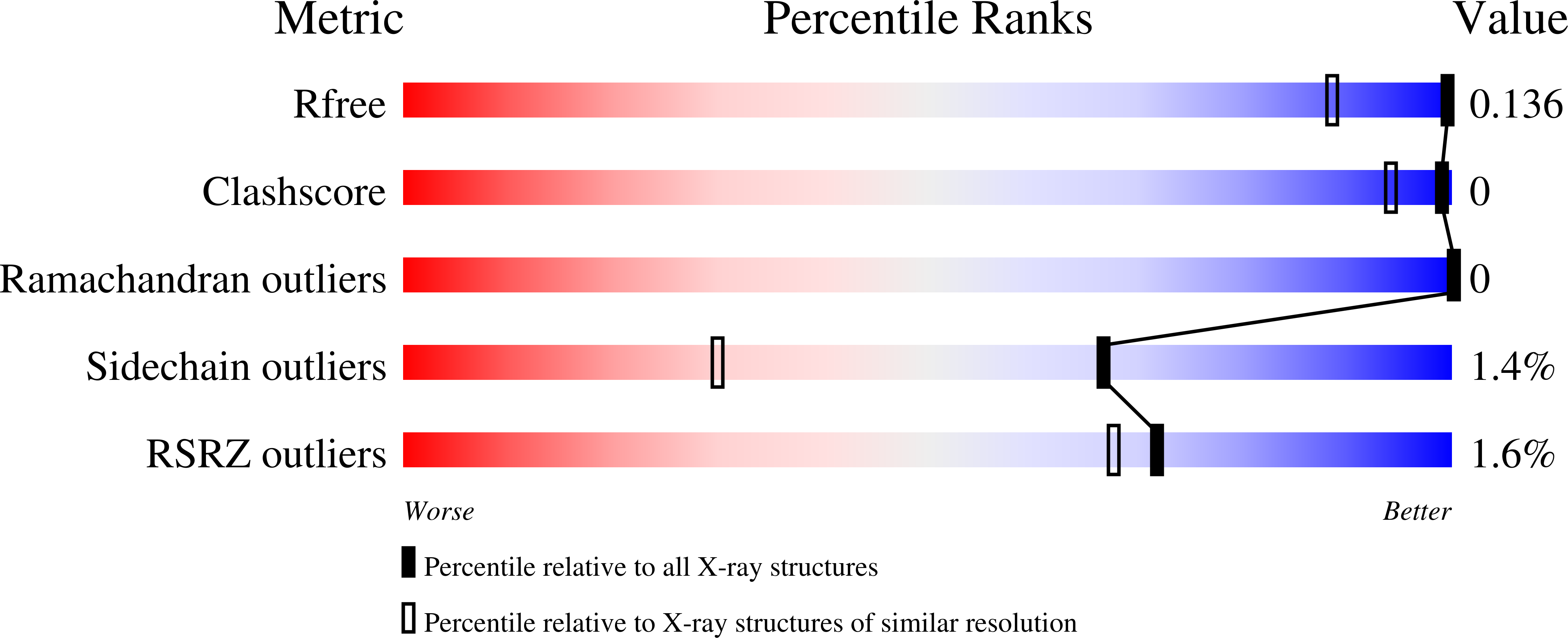
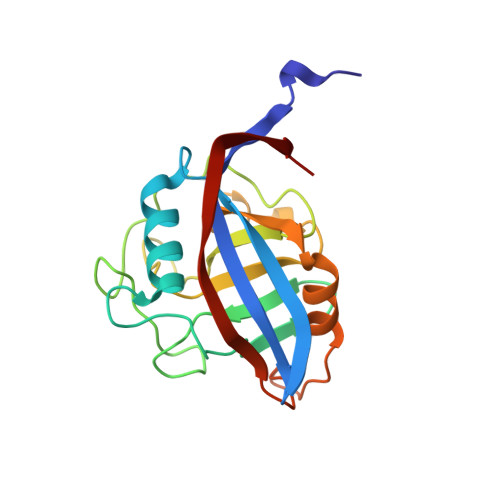
Crystal structures of wild-type and mutated cyclophilin B that causes hyperelastosis cutis in the American quarter horse.
Boudko, S.P., Ishikawa, Y., Lerch, T.F., Nix, J., Chapman, M.S., Bachinger, H.P.(2012) BMC Res Notes 5: 626-626
- PubMed: 23137129
- DOI: https://doi.org/10.1186/1756-0500-5-626
- Primary Citation of Related Structures:
4FRU, 4FRV - PubMed Abstract:
Hyperelastosis cutis is an inherited autosomal recessive connective tissue disorder. Affected horses are characterized by hyperextensible skin, scarring, and severe lesions along the back. The disorder is caused by a mutation in cyclophilin B. The crystal structures of both wild-type and mutated (Gly6->Arg) horse cyclophilin B are presented. The mutation neither affects the overall fold of the enzyme nor impairs the catalytic site structure. Instead, it locally rearranges the flexible N-terminal end of the polypeptide chain and also makes it more rigid. Interactions of the mutated cyclophilin B with a set of endoplasmic reticulum-resident proteins must be affected.
Organizational Affiliation:
Research Department, Shriners Hospital for Children, Portland, OR 97239, USA.