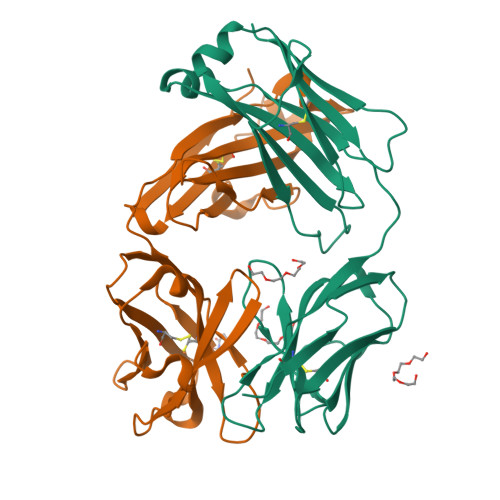
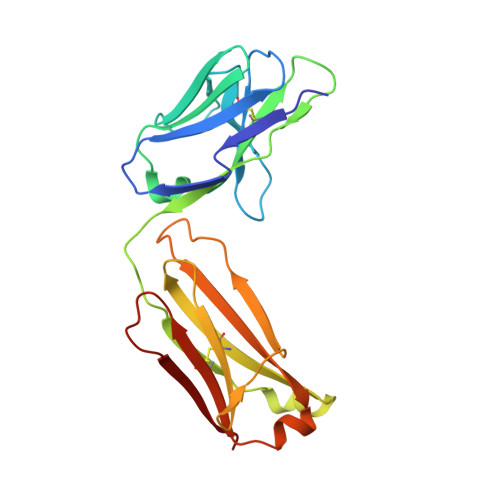
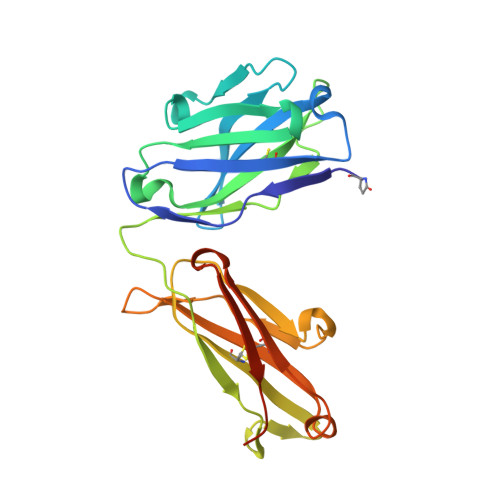
Recognition of mesothelin by the therapeutic antibody MORAb-009: structural and mechanistic insights.
Ma, J., Tang, W.K., Esser, L., Pastan, I., Xia, D.(2012) J Biological Chem 287: 33123-33131
- PubMed: 22787150
- DOI: https://doi.org/10.1074/jbc.M112.381756
- Primary Citation of Related Structures:
4F33, 4F3F - PubMed Abstract:
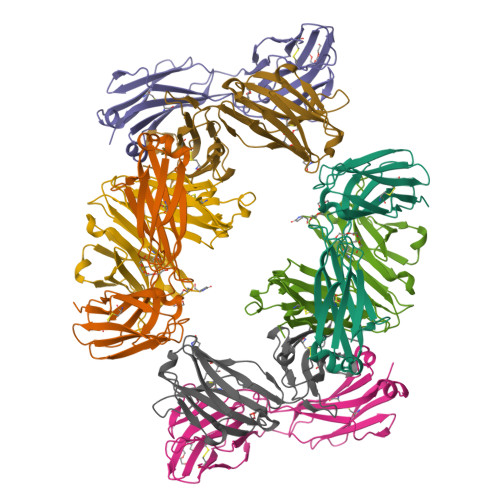
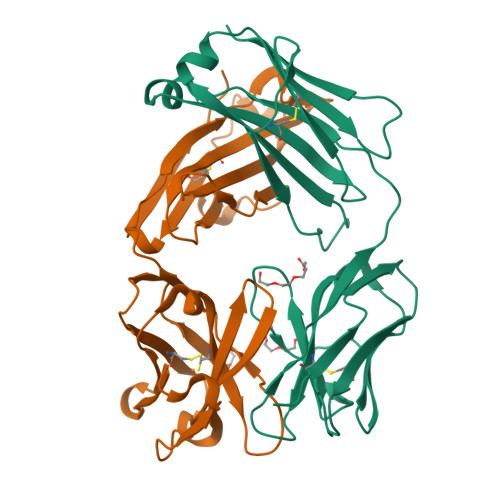
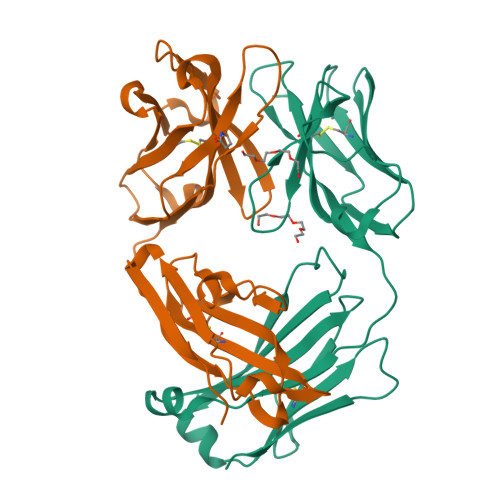
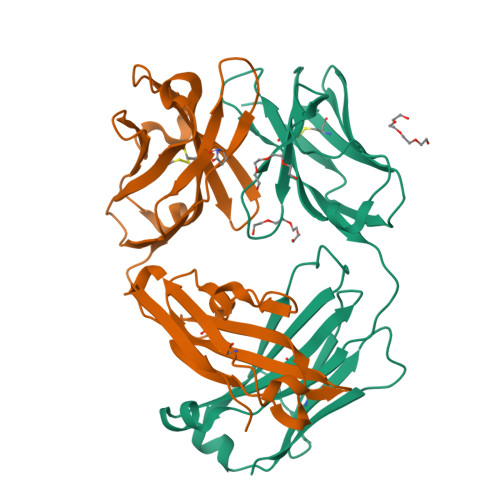
Mesothelin is a tumor differentiation antigen that is highly expressed in many epithelial cancers, with limited expression in normal human tissues. Binding of mesothelin on normal mesothelial cells lining the pleura or peritoneum to the tumor-associated cancer antigen 125 (CA-125) can lead to heterotypic cell adhesion and tumor metastasis within the pleural and peritoneal cavities. This binding can be prevented by MORAb-009, a humanized monoclonal antibody against mesothelin currently under clinical trials. We show here that MORAb-009 recognizes a non-linear epitope that is contained in the first 64-residue fragment of the mesothelin. We further demonstrate that the recognition is independent of glycosylation state of the protein but sensitive to the loss of a disulfide bond linking residues Cys-7 and Cys-31. The crystal structure of the complex between the mesothelin N-terminal fragment and Fab of MORAb-009 at 2.6 Å resolution reveals an epitope encompassing multiple secondary structural elements of the mesothelin, including residues from helix α1, the loops linking helices α1 and α2, and between helices α4 and α5. The mesothelin fragment has a compact, right-handed superhelix structure consisting of five short helices and connecting loops. A residue essential for complex formation has been identified as Phe-22, which projects its side chain into a hydrophobic niche formed on the antibody recognition surface upon antigen-antibody contact. The overlapping binding footprints of both the monoclonal antibody and the cancer antigen CA-125 explains the therapeutic effect and provides a basis for further antibody improvement.
Organizational Affiliation:
Laboratory of Cell Biology, Center for Cancer Research, NCI, National Institutes of Health, Bethesda, Maryland 20892, USA.