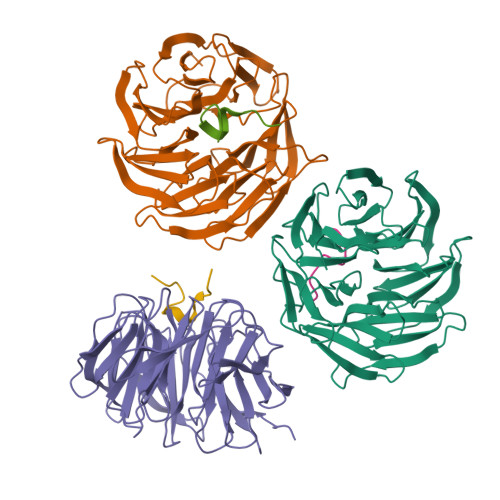
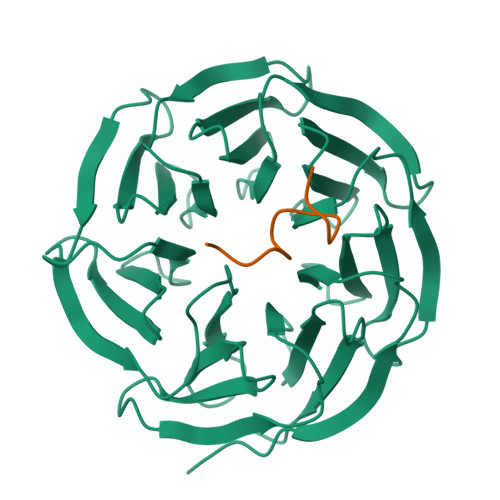
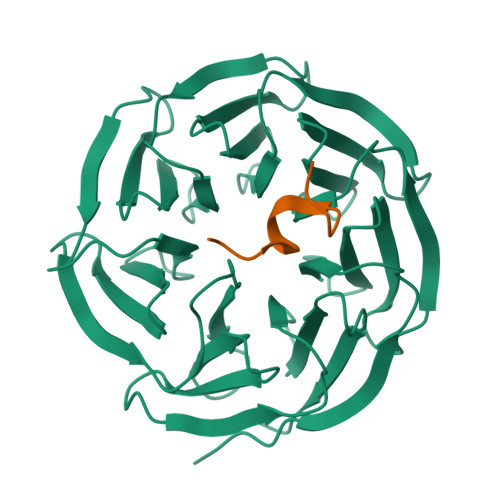
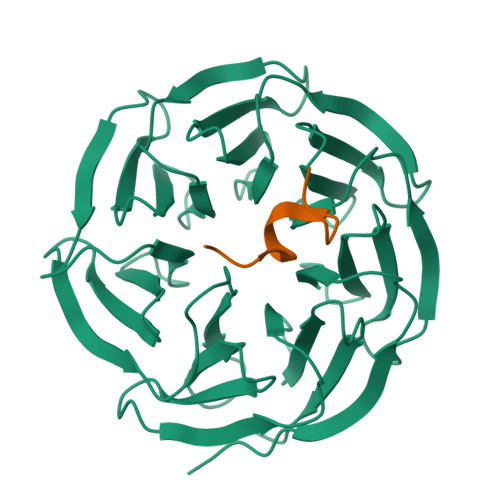
Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
Dharmarajan, V., Lee, J.H., Patel, A., Skalnik, D.G., Cosgrove, M.S.(2012) J Biological Chem 287: 27275-27289
- PubMed: 22665483
- DOI: https://doi.org/10.1074/jbc.M112.364125
- Primary Citation of Related Structures:
4ERQ, 4ERY, 4ERZ, 4ES0, 4ESG, 4EWR - PubMed Abstract:
Translocations and amplifications of the mixed lineage leukemia-1 (MLL1) gene are associated with aggressive myeloid and lymphocytic leukemias in humans. MLL1 is a member of the SET1 family of histone H3 lysine 4 (H3K4) methyltransferases, which are required for transcription of genes involved in hematopoiesis and development. MLL1 associates with a subcomplex containing WDR5, RbBP5, Ash2L, and DPY-30 (WRAD), which together form the MLL1 core complex that is required for sequential mono- and dimethylation of H3K4. We previously demonstrated that WDR5 binds the conserved WDR5 interaction (Win) motif of MLL1 in vitro, an interaction that is required for the H3K4 dimethylation activity of the MLL1 core complex. In this investigation, we demonstrate that arginine 3765 of the MLL1 Win motif is required to co-immunoprecipitate WRAD from mammalian cells, suggesting that the WDR5-Win motif interaction is important for the assembly of the MLL1 core complex in vivo. We also demonstrate that peptides that mimic SET1 family Win motif sequences inhibit H3K4 dimethylation by the MLL1 core complex with varying degrees of efficiency. To understand the structural basis for these differences, we determined structures of WDR5 bound to six different naturally occurring Win motif sequences at resolutions ranging from 1.9 to 1.2 Å. Our results reveal that binding energy differences result from interactions between non-conserved residues C-terminal to the Win motif and to a lesser extent from subtle variation of residues within the Win motif. These results highlight a new class of methylation inhibitors that may be useful for the treatment of MLL1-related malignancies.
Organizational Affiliation:
Department of Biology, Syracuse University, Syracuse, New York 13244, USA.