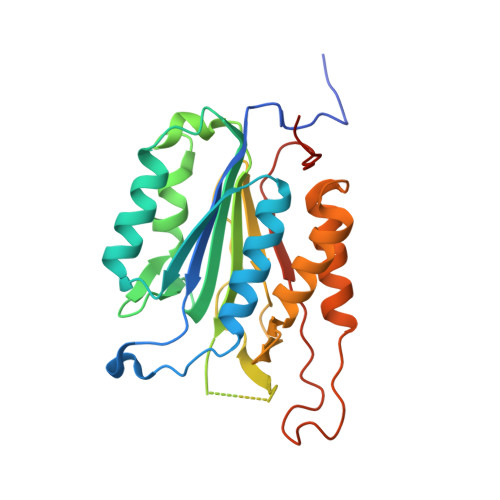
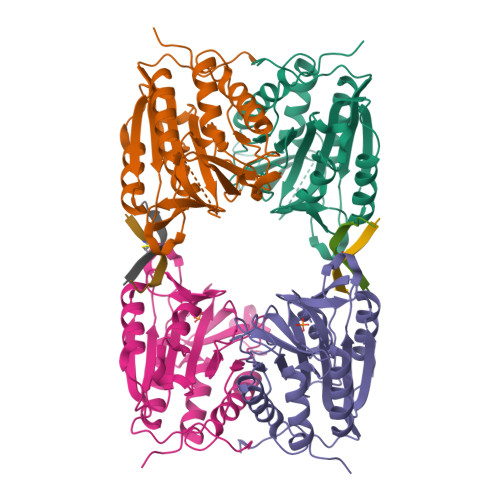Allosteric peptides bind a caspase zymogen and mediate caspase tetramerization.
Stanger, K., Steffek, M., Zhou, L., Pozniak, C.D., Quan, C., Franke, Y., Tom, J., Tam, C., Elliott, J.M., Lewcock, J.W., Zhang, Y., Murray, J., Hannoush, R.N.(2012) Nat Chem Biol 8: 655-660
- PubMed: 22683611
- DOI: https://doi.org/10.1038/nchembio.967
- Primary Citation of Related Structures:
4EJF - PubMed Abstract:
The caspases are a family of cytosolic proteases with essential roles in inflammation and apoptosis. Drug discovery efforts have focused on developing molecules directed against the active sites of caspases, but this approach has proved challenging and has not yielded any approved therapeutics. Here we describe a new strategy for generating inhibitors of caspase-6, a potential therapeutic target in neurodegenerative disorders, by screening against its zymogen form. Using phage display to discover molecules that bind the zymogen, we report the identification of a peptide that specifically impairs the function of caspase-6 in vitro and in neuronal cells. Remarkably, the peptide binds at a tetramerization interface that is uniquely present in zymogen caspase-6, rather than binding into the active site, and acts via a new allosteric mechanism that promotes caspase tetramerization. Our data illustrate that screening against the zymogen holds promise as an approach for targeting caspases in drug discovery.
Organizational Affiliation:
Department of Early Discovery Biochemistry, Genentech, South San Francisco, California, USA.