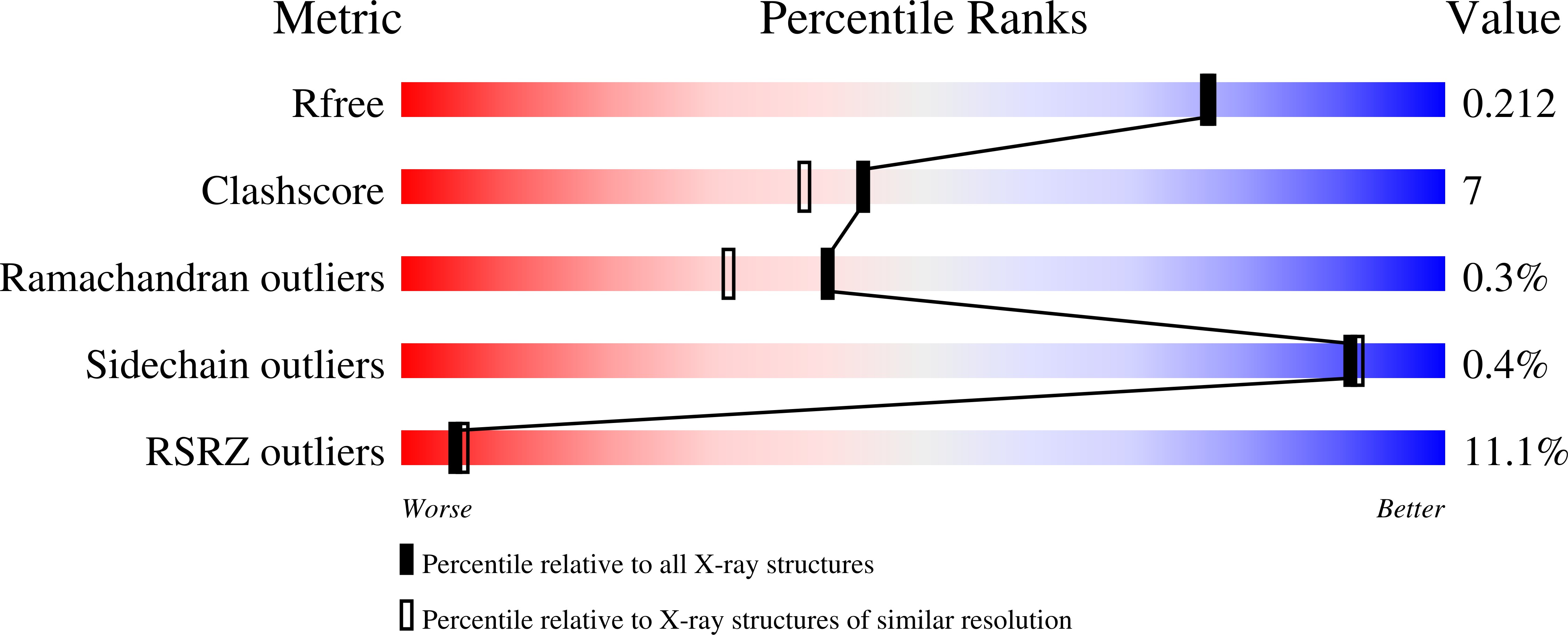
Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Emptage, R.P., Daughtry, K.D., Pemble, C.W., Raetz, C.R.(2012) Proc Natl Acad Sci U S A 109: 12956-12961
- PubMed: 22826246
- DOI: https://doi.org/10.1073/pnas.1206072109
- Primary Citation of Related Structures:
4EHW, 4EHX, 4EHY - PubMed Abstract:
In Gram-negative bacteria, the hydrophobic anchor of the outer membrane lipopolysaccharide is lipid A, a saccharolipid that plays key roles in both viability and pathogenicity of these organisms. The tetraacyldisaccharide 4'-kinase (LpxK) of the diverse P-loop-containing nucleoside triphosphate hydrolase superfamily catalyzes the sixth step in the biosynthetic pathway of lipid A, and is the only known P-loop kinase to act upon a lipid substrate at the membrane. Here, we report the crystal structures of apo- and ADP/Mg(2+)-bound forms of Aquifex aeolicus LpxK to a resolution of 1.9 Å and 2.2 Å, respectively. LpxK consists of two α/β/α sandwich domains connected by a two-stranded β-sheet linker. The N-terminal domain, which has most structural homology to other family members, is responsible for catalysis at the P-loop and positioning of the disaccharide-1-phosphate substrate for phosphoryl transfer on the inner membrane. The smaller C-terminal domain, a substructure unique to LpxK, helps bind the nucleotide substrate and Mg(2+) cation using a 25° hinge motion about its base. Activity was severely reduced in alanine point mutants of conserved residues D138 and D139, which are not directly involved in ADP or Mg(2+) binding in our structures, indicating possible roles in phosphoryl acceptor positioning or catalysis. Combined structural and kinetic studies have led to an increased understanding of the enzymatic mechanism of LpxK and provided the framework for structure-based antimicrobial design.
Organizational Affiliation:
Department of Biochemistry and Human Vaccine Institute, Duke University Medical Center, Durham, NC 27710, USA.