Pyrazole and pyrimidine phenylacylsulfonamides as dual Bcl-2/Bcl-xL antagonists.
Schroeder, G.M., Wei, D., Banfi, P., Cai, Z.W., Lippy, J., Menichincheri, M., Modugno, M., Naglich, J., Penhallow, B., Perez, H.L., Sack, J., Schmidt, R.J., Tebben, A., Yan, C., Zhang, L., Galvani, A., Lombardo, L.J., Borzilleri, R.M.(2012) Bioorg Med Chem Lett 22: 3951-3956
- PubMed: 22608393
- DOI: https://doi.org/10.1016/j.bmcl.2012.04.106
- Primary Citation of Related Structures:
4EHR - PubMed Abstract:
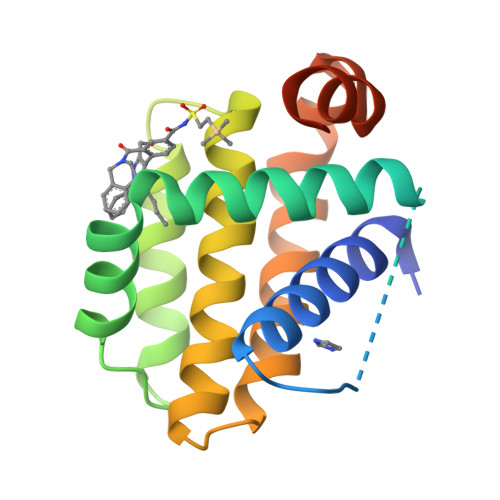
5-Butyl-1,4-diphenyl pyrazole and 2-amino-5-chloro pyrimidine acylsulfonamides were developed as potent dual antagonists of Bcl-2 and Bcl-xL. Compounds were optimized for binding to the I88, L92, I95, and F99 pockets normally occupied by pro-apoptotic protein Bim. An X-ray crystal structure confirmed the proposed binding mode. Observation of cytochrome c release from isolated mitochondria in MV-411 cells provides further evidence of target inhibition. Compounds demonstrated submicromolar antiproliferative activity in Bcl-2/Bcl-xL dependent cell lines.
Organizational Affiliation:
Bristol-Myers Squibb Research, PO Box 4000, Princeton, NJ 08543-4000, USA. gretchen.schroeder@bms.com