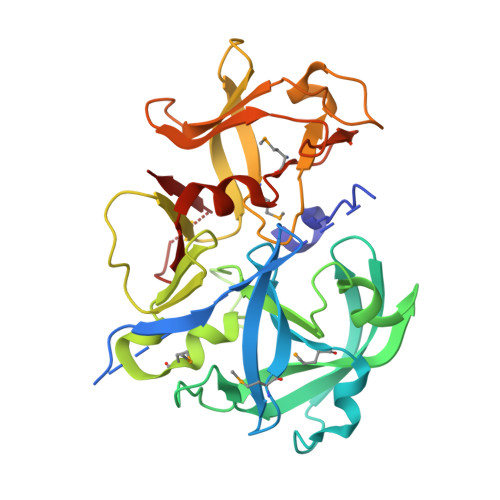
Crystal structure of a putative aspartic proteinase domain of the Mycobacterium tuberculosis cell surface antigen PE_PGRS16
Barathy, D.V., Suguna, K.(2013) FEBS Open Bio 3: 256-262
- PubMed: 23923105
- DOI: https://doi.org/10.1016/j.fob.2013.05.004
- Primary Citation of Related Structures:
4EHC - PubMed Abstract:
We report the crystal structure of the first prokaryotic aspartic proteinase-like domain identified in the genome of Mycobacterium tuberculosis. A search in the genomes of Mycobacterium species showed that the C-terminal domains of some of the PE family proteins contain two classic DT/SG motifs of aspartic proteinases with a low overall sequence similarity to HIV proteinase. The three-dimensional structure of one of them, Rv0977 (PE_PGRS16) of M. tuberculosis revealed the characteristic pepsin-fold and catalytic site architecture. However, the active site was completely blocked by the N-terminal His-tag. Surprisingly, the enzyme was found to be inactive even after the removal of the N-terminal His-tag. A comparison of the structure with pepsins showed significant differences in the critical substrate binding residues and in the flap tyrosine conformation that could contribute to the lack of proteolytic activity of Rv0977.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India.