Insight into structural diversity of influenza virus haemagglutinin
Cho, K.J., Lee, J.H., Hong, K.W., Kim, S.H., Park, Y., Lee, J.Y., Kang, S., Kim, S., Yang, J.H., Kim, E.K., Seok, J.H., Unzai, S., Park, S.Y., Saelens, X., Kim, C.J., Lee, J.Y., Kang, C., Oh, H.B., Chung, M.S., Kim, K.H.(2013) J Gen Virol 94: 1712-1722
- PubMed: 23636824
- DOI: https://doi.org/10.1099/vir.0.051136-0
- Primary Citation of Related Structures:
4EDA, 4EDB - PubMed Abstract:
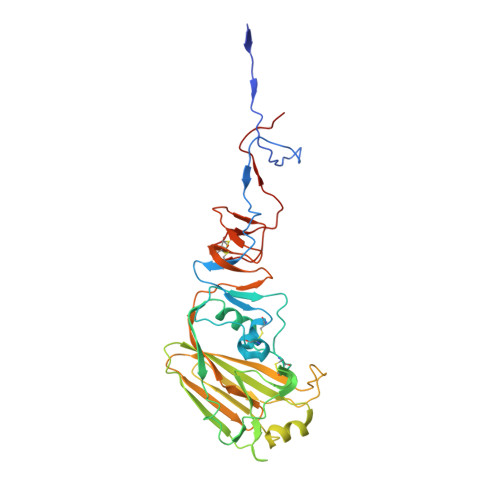
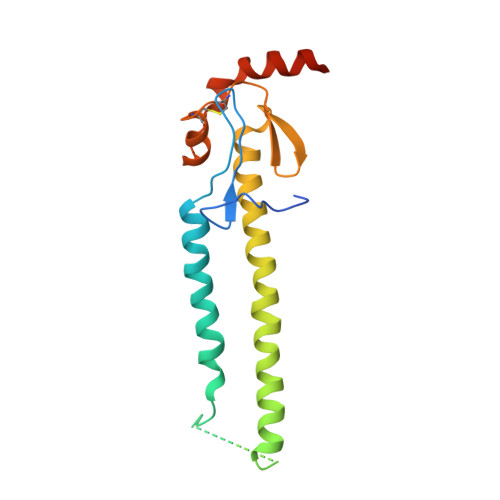
Influenza virus infects host cells through membrane fusion, a process mediated by the low pH-induced conformational change of the viral surface glycoprotein haemagglutinin (HA). We determined the structures and biochemical properties of the HA proteins from A/Korea/01/2009 (KR01), a 2009 pandemic strain, and A/Thailand/CU44/2006 (CU44), a seasonal strain. The crystal structure of KR01 HA revealed a V-shaped head-to-head arrangement, which is not seen in other HA proteins including CU44 HA. We isolated a broadly neutralizing H1-specific monoclonal antibody GC0757. The KR01 HA-Fab0757 complex structure also exhibited a head-to-head arrangement of HA. Both native and Fab complex structures reveal a different spatial orientation of HA1 relative to HA2, indicating that HA is flexible and dynamic at neutral pH. Further, the KR01 HA exhibited significantly lower protein stability and increased susceptibility to proteolytic cleavage compared with other HAs. Our structures provide important insights into the conformational flexibility of HA.
Organizational Affiliation:
Department of Biotechnology & Bioinformatics, College of Science & Technology, Korea University, Sejong 339-700, Korea.