Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
Zhang, J., Chen, Y., Qi, J., Gao, F., Liu, Y., Liu, J., Zhou, X., Kaufman, J., Xia, C., Gao, G.F.(2012) J Immunol 189: 4478-4487
- PubMed: 23041567
- DOI: https://doi.org/10.4049/jimmunol.1200885
- Primary Citation of Related Structures:
4E0R, 4G42, 4G43 - PubMed Abstract:
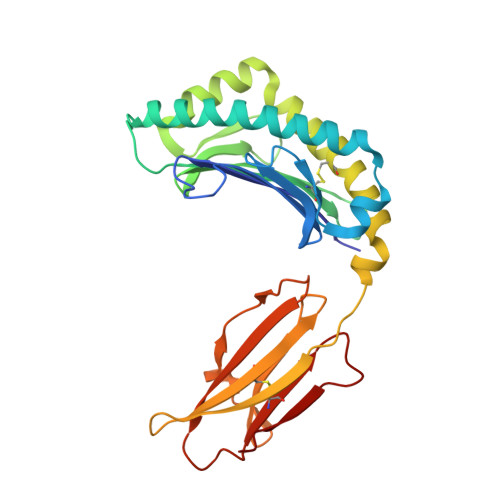
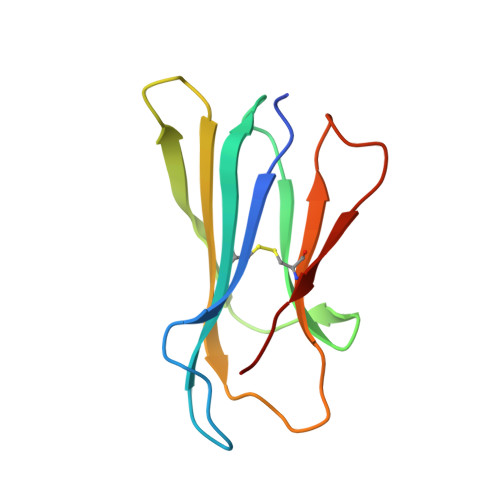
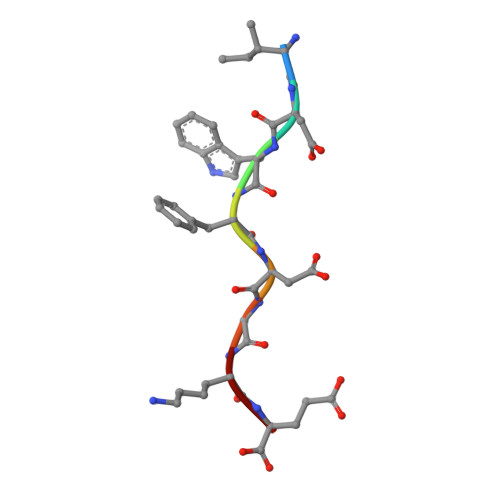
The MHC has genetic associations with many diseases, often due to differences in presentation of antigenic peptides by polymorphic MHC molecules to T lymphocytes of the immune system. In chickens, only a single classical class I molecule in each MHC haplotype is expressed well due to coevolution with the polymorphic TAPs which means that resistance and susceptibility to infectious pathogens are particularly easy to observe. Previously, structures of chicken MHC class I molecule BF2*2101 from B21 haplotype showed an unusually large peptide-binding groove that accommodates a broad spectrum of peptides to present as epitopes to CTLs, explaining the MHC-determined resistance of B21 chickens to Marek's disease. In this study, we report the crystal structure of BF2*0401 from the B4 (also known as B13) haplotype, showing a highly positively charged surface hitherto unobserved in other MHC molecules, as well as a remarkably narrow groove due to the allele-specific residues with bulky side chains. Together, these properties limit the number of epitope peptides that can bind this class I molecule. However, peptide-binding assays show that in vitro, BF2*0401 can bind a wider variety of peptides than are found on the surface of B4 cells. Thus, a combination of the specificities of the polymorphic TAP and the MHC results in a very limited set of BF2*0401 peptides with negatively charged anchors to be presented to T lymphocytes.
Organizational Affiliation:
Department of Microbiology and Immunology, College of Veterinary Medicine, China Agricultural University, Beijing 100094, China.