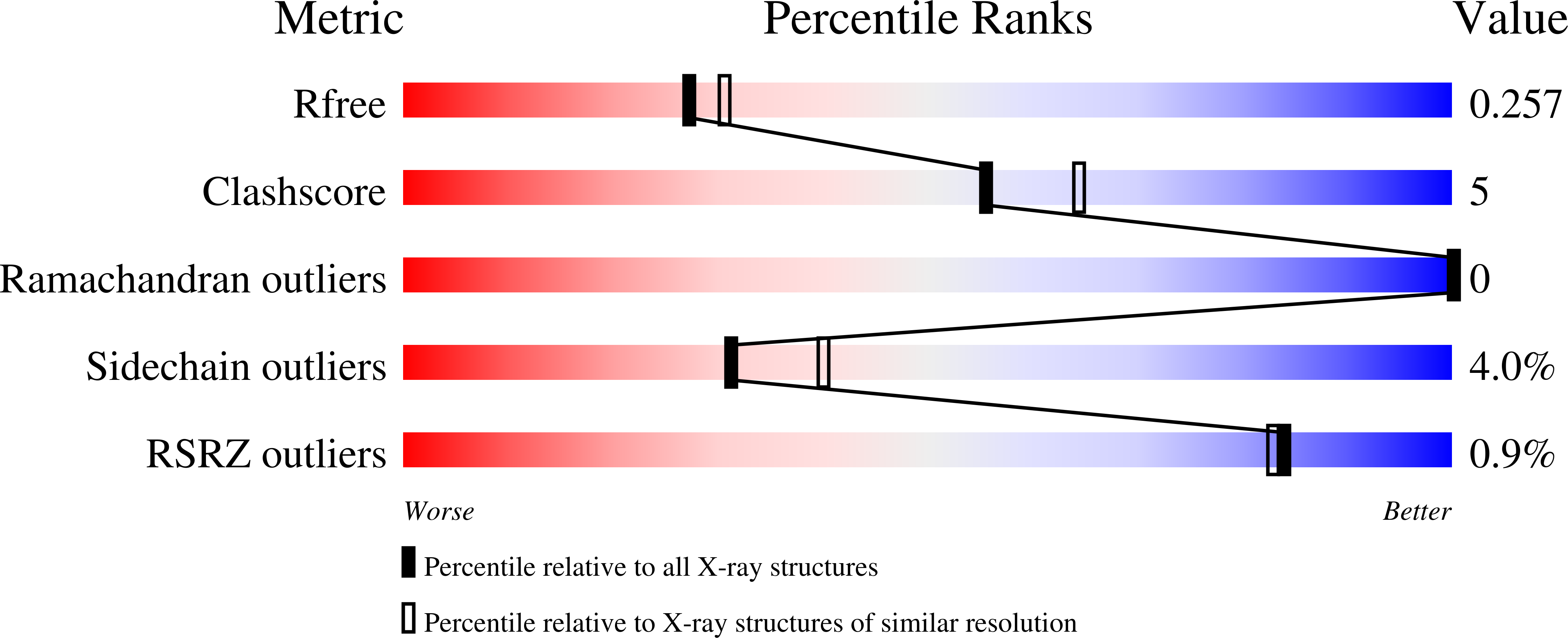
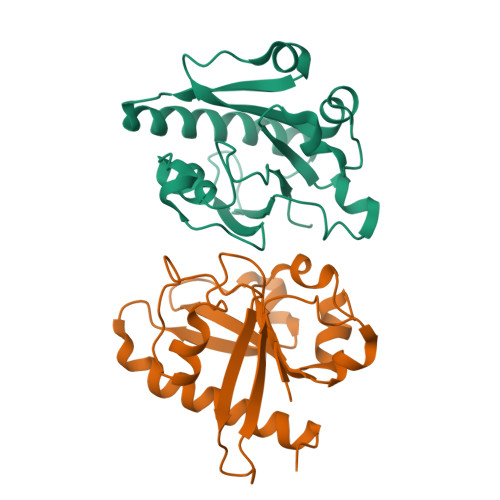
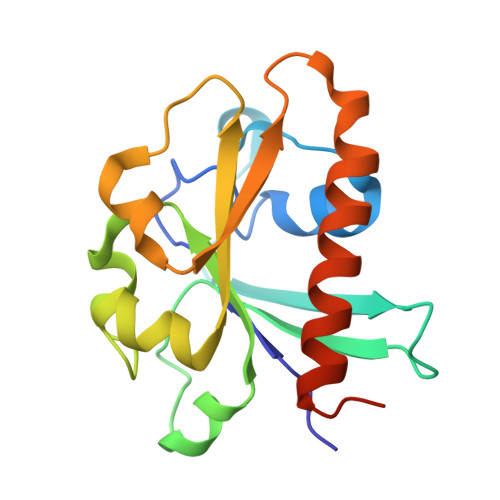
Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold.
Chen, H., Shi, N., Gao, Y., Li, X., Teng, M., Niu, L.(2012) Acta Crystallogr D Biol Crystallogr 68: 953-959
- PubMed: 22868760
- DOI: https://doi.org/10.1107/S0907444912017325
- Primary Citation of Related Structures:
4DM4 - PubMed Abstract:
The yeast Paf1 complex (Paf1C), which is composed of the proteins Paf1, Cdc73, Ctr9, Leo1 and Rtf1, accompanies RNA polymerase II from the promoter to the 3'-end formation site of mRNA- and snoRNA-encoding genes. As one of the first identified subunits of Paf1C, yeast Cdc73 (yCdc73) takes part in many transcription-related processes, including binding to RNA polymerase II, recruitment and activation of histone-modification factors and communication with other transcriptional activators. The human homologue of yCdc73, parafibromin, has been identified as a tumour suppressor linked to breast, renal and gastric cancers. However, the functional mechanism of yCdc73 has until recently been unclear. Here, a 2.2 Å resolution crystal structure of the highly conserved C-terminal region of yCdc73 is reported. It revealed that yCdc73 appears to have a GTPase-like fold. However, no GTPase activity was observed. The crystal structure of yCdc73 will shed new light on the modes of function of Cdc73 and Paf1C.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, People's Republic of China.