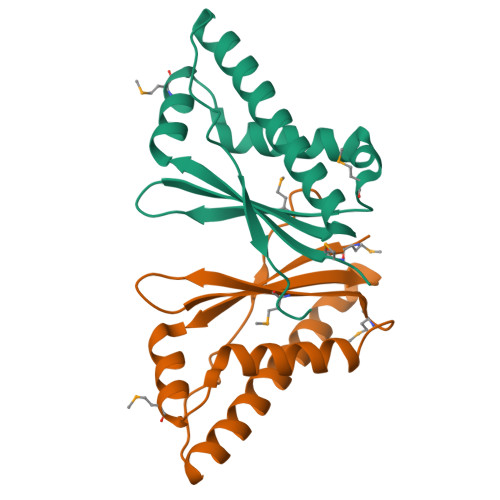
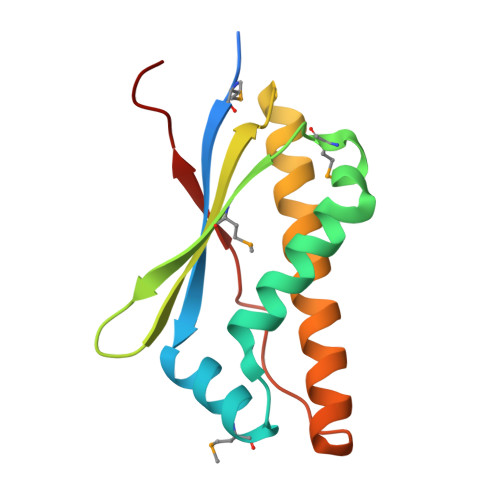
A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors.
Ou, H.D., Kwiatkowski, W., Deerinck, T.J., Noske, A., Blain, K.Y., Land, H.S., Soria, C., Powers, C.J., May, A.P., Shu, X., Tsien, R.Y., Fitzpatrick, J.A., Long, J.A., Ellisman, M.H., Choe, S., O'Shea, C.C.(2012) Cell 151: 304-319
- PubMed: 23063122
- DOI: https://doi.org/10.1016/j.cell.2012.08.035
- Primary Citation of Related Structures:
4DJB - PubMed Abstract:
Evolution of minimal DNA tumor virus' genomes has selected for small viral oncoproteins that hijack critical cellular protein interaction networks. The structural basis for the multiple and dominant functions of adenovirus oncoproteins has remained elusive. E4-ORF3 forms a nuclear polymer and simultaneously inactivates p53, PML, TRIM24, and MRE11/RAD50/NBS1 (MRN) tumor suppressors. We identify oligomerization mutants and solve the crystal structure of E4-ORF3. E4-ORF3 forms a dimer with a central β core, and its structure is unrelated to known polymers or oncogenes. E4-ORF3 dimer units coassemble through reciprocal and nonreciprocal exchanges of their C-terminal tails. This results in linear and branched oligomer chains that further assemble in variable arrangements to form a polymer network that partitions the nuclear volume. E4-ORF3 assembly creates avidity-driven interactions with PML and an emergent MRN binding interface. This reveals an elegant structural solution whereby a small protein forms a multivalent matrix that traps disparate tumor suppressors.
Organizational Affiliation:
Molecular and Cell Biology Laboratory, Salk Institute for Biological Studies, 10010 North Torrey Pines Road, La Jolla, CA 92037, USA.