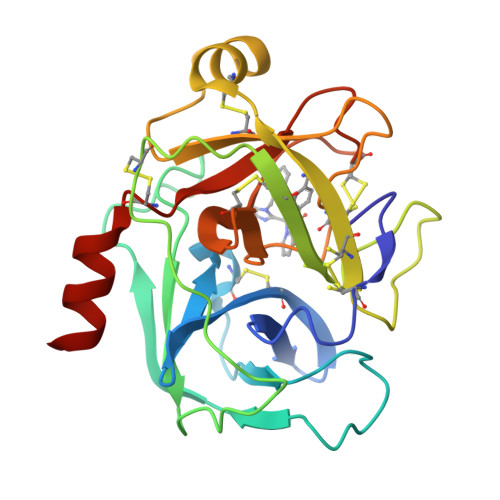
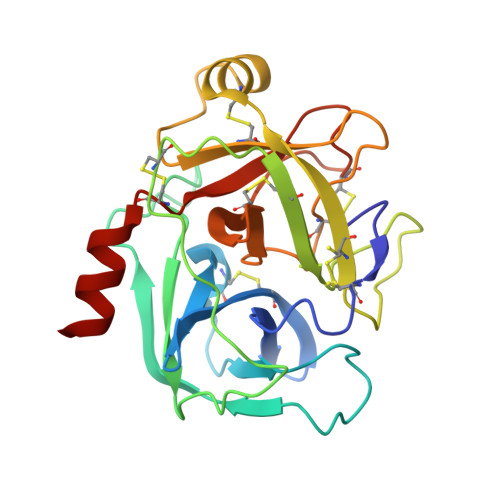
Human kallikrein 6 inhibitors with a para-amidobenzylanmine P1 group identified through virtual screening.
Liang, G., Chen, X., Aldous, S., Pu, S.F., Mehdi, S., Powers, E., Xia, T., Wang, R.(2012) Bioorg Med Chem Lett 22: 2450-2455
- PubMed: 22386244
- DOI: https://doi.org/10.1016/j.bmcl.2012.02.014
- Primary Citation of Related Structures:
4D8N - PubMed Abstract:
A series of hK6 inhibitors with a para-amidobenzylamine P1 group and a 2-hydroxybenzamide scaffold linker was discovered through virtual screening. The X-ray structure of hK6 complexed with compound 9b was determined to a resolution of 1.68Å. The tertiary folding of the hK6 complexed with the inhibitor is conserved relative to the structure of the apo-protein, whereas the interaction between hK6 and the inhibitor is consistent with both the SAR and the in silico model used in the virtual screening.
Organizational Affiliation:
Molecular Innovative Therapeutics, Sanofi Pharmaceuticals, Bridgewater, NJ, USA. guyan.liang@verizon.net