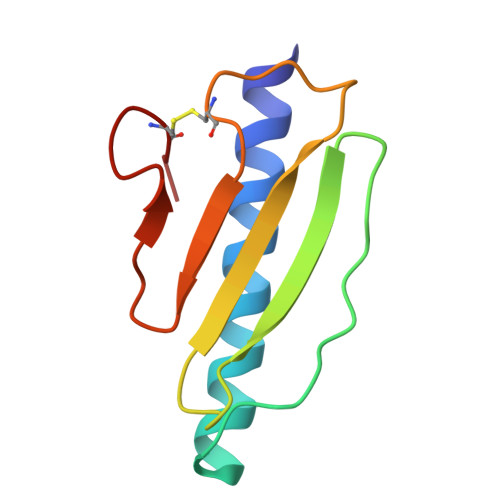
High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Gorgel, M., Ulstrup, J.J., Boggild, A., Jones, N.C., Hoffmann, S.V., Nissen, P., Boesen, T.(2015) BMC Struct Biol 15: 4
- PubMed: 25886849
- DOI: https://doi.org/10.1186/s12900-015-0031-7
- Primary Citation of Related Structures:
4D40, 4US7 - PubMed Abstract:
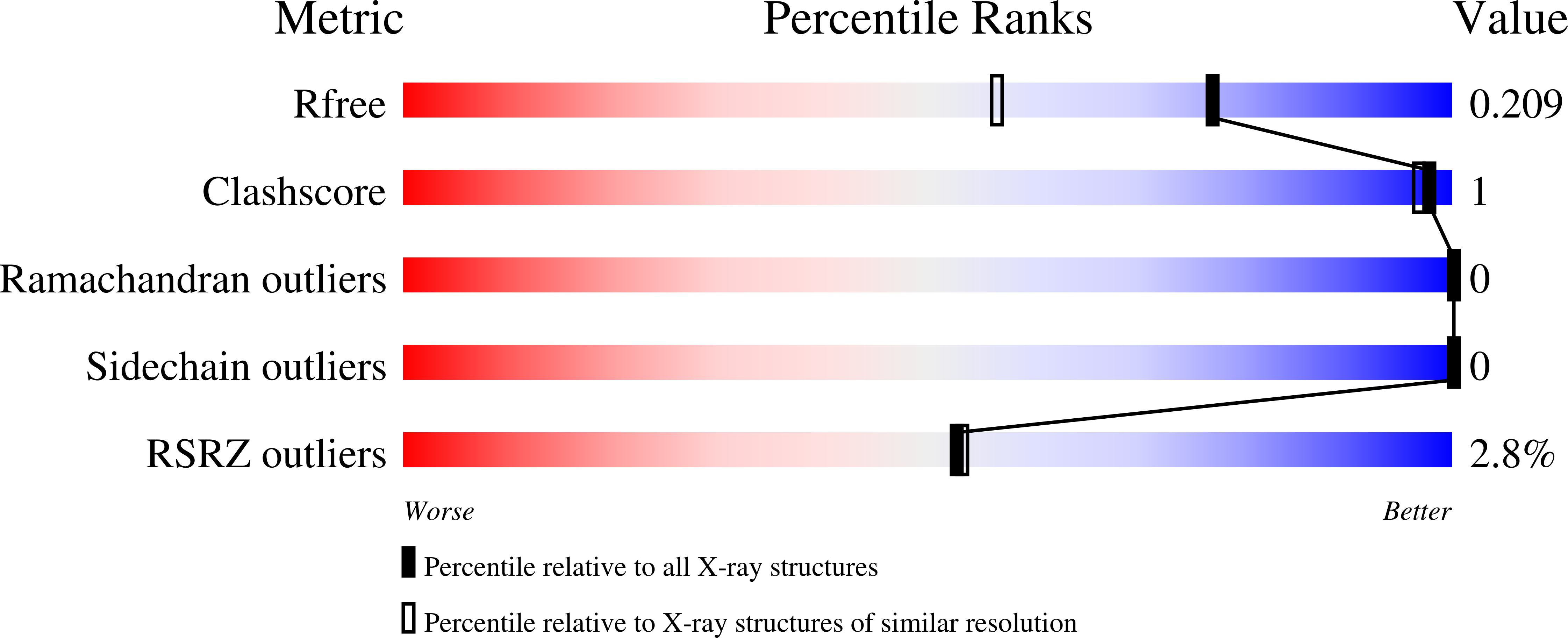
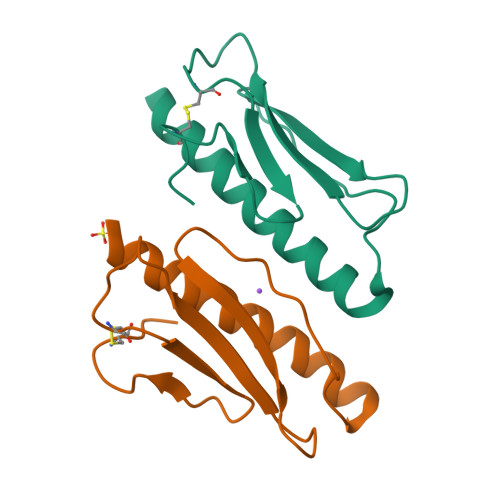
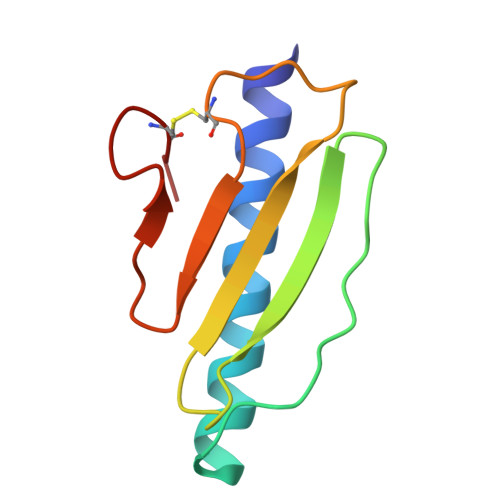
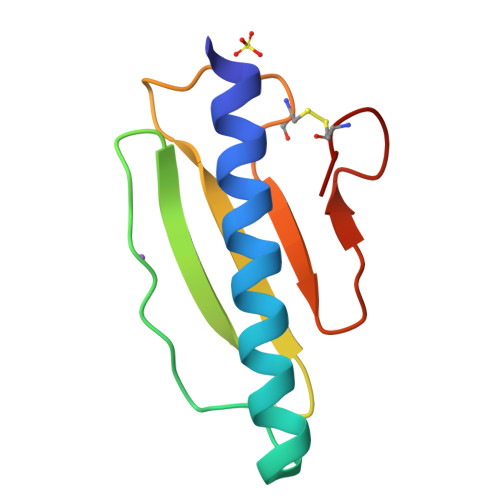
Type IV pili are widely expressed among Gram-negative bacteria, where they are involved in biofilm formation, serve in the transfer of DNA, motility and in the bacterial attachment to various surfaces. Type IV pili in Shewanella oneidensis are also supposed to play an important role in extracellular electron transfer by the attachment to sediments containing electron acceptors and potentially forming conductive nanowires. The potential nanowire type IV pilin PilBac1 from S. oneidensis was characterized by a combination of complementary structural methods and the atomic structure was determined at a resolution of 1.67 Å by X-ray crystallography. PilBac1 consists of one long N-terminal α-helix packed against four antiparallel β-strands, thus revealing the core fold of type IV pilins. In the crystal, PilBac1 forms a parallel dimer with a sodium ion bound to one of the monomers. Interestingly, our PilBac1 crystal structure reveals two unusual features compared to other type IVa pilins: an unusual position of the disulfide bridge and a straight α-helical section, which usually exhibits a pronounced kink. This straight helix leads to a distinct packing in a filament model of PilBac1 based on an EM model of a Neisseria pilus. In this study we have described the first structure of a pilin from Shewanella oneidensis. The structure possesses features of the common type IV pilin core, but also exhibits significant variations in the α-helical part and the D-region.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 10c, Aarhus C, 8000, Denmark. manuela@mbg.au.dk.