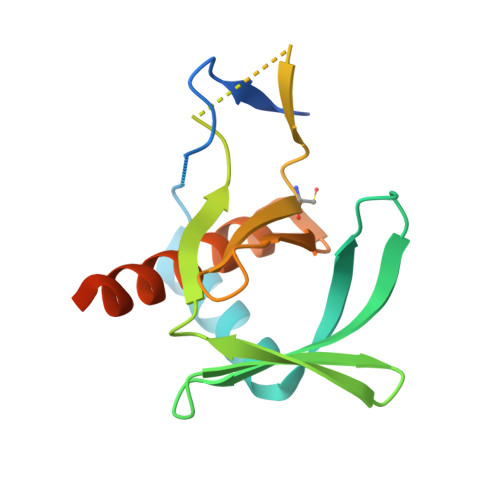
The Inner Membrane Complex Sub-Compartment Proteins Critical for Replication of the Apicomplexan Parasite Toxoplasma Gondii Adopt a Pleckstrin Homology Fold
Tonkin, M.L., Beck, J.R., Bradley, P.J., Boulanger, M.J.(2014) J Biological Chem 289: 13962
- PubMed: 24675080
- DOI: https://doi.org/10.1074/jbc.M114.548891
- Primary Citation of Related Structures:
4CHJ, 4CHM - PubMed Abstract:
Toxoplasma gondii, an apicomplexan parasite prevalent in developed nations, infects up to one-third of the human population. The success of this parasite depends on several unique structures including an inner membrane complex (IMC) that lines the interior of the plasma membrane and contains proteins important for gliding motility and replication. Of these proteins, the IMC sub-compartment proteins (ISPs) have recently been shown to play a role in asexual T. gondii daughter cell formation, yet the mechanism is unknown. Complicating mechanistic characterization of the ISPs is a lack of sequence identity with proteins of known structure or function. In support of elucidating the function of ISPs, we first determined the crystal structures of representative members TgISP1 and TgISP3 to a resolution of 2.10 and 2.32 Å, respectively. Structural analysis revealed that both ISPs adopt a pleckstrin homology fold often associated with phospholipid binding or protein-protein interactions. Substitution of basic for hydrophobic residues in the region that overlays with phospholipid binding in related pleckstrin homology domains, however, suggests that ISPs do not retain phospholipid binding activity. Consistent with this observation, biochemical assays revealed no phospholipid binding activity. Interestingly, mapping of conserved surface residues combined with crystal packing analysis indicates that TgISPs have functionally repurposed the phospholipid-binding site likely to coordinate protein partners. Recruitment of larger protein complexes may also be aided through avidity-enhanced interactions resulting from multimerization of the ISPs. Overall, we propose a model where TgISPs recruit protein partners to the IMC to ensure correct progression of daughter cell formation.
Organizational Affiliation:
From the Department of Biochemistry and Microbiology, University of Victoria, Victoria, British Columbia V8W 3P6, Canada and.