Two High-Resolution Structures of the Human E3 Ubiquitin Ligase Siah1.
Rimsa, V., Eadsforth, T.C., Hunter, W.N.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 96: 1339
- PubMed: 24316825
- DOI: https://doi.org/10.1107/S1744309113031448
- Primary Citation of Related Structures:
4C9Z, 4CA1 - PubMed Abstract:
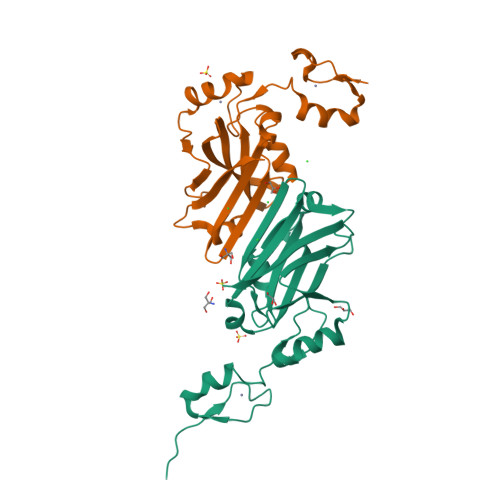
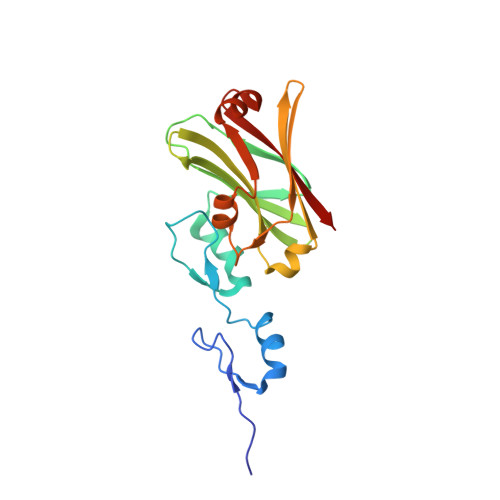
Siah1 is an E3 ubiquitin ligase that contributes to proteasome-mediated degradation of multiple targets in key cellular processes and which shows promise as a therapeutic target in oncology. Structures of a truncated Siah1 bound to peptide-based inhibitors have been reported. Here, new crystallization conditions have allowed the determination of a construct encompassing dual zinc-finger subdomains and substrate-binding domains at significantly higher resolution. Although the crystals appear isomorphous, two structures present distinct states in which the spatial orientation of one zinc-finger subdomain differs with respect to the rest of the dimeric protein. Such a difference, which is indicative of conformational freedom, infers potential biological relevance related to recognition of binding partners. The crystallization conditions and improved models of Siah1 may aid future studies investigating Siah1-ligand complexes.
Organizational Affiliation:
Division of Biological Chemistry and Drug Discovery, College of Life Sciences, University of Dundee, Dundee DD1 5EH, Scotland.