Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Zebisch, M., Xu, Y., Krastev, C., Macdonald, B.T., Chen, M., Gilbert, R.J.C., He, X., Jones, E.Y.(2013) Nat Commun 4: 2787
- PubMed: 24225776
- DOI: https://doi.org/10.1038/ncomms3787
- Primary Citation of Related Structures:
4C84, 4C85, 4C86, 4C8A, 4C8C, 4C8F, 4C8P, 4C8T, 4C8U, 4C8V, 4C8W, 4C99, 4C9A, 4C9E, 4C9R, 4C9U, 4C9V - PubMed Abstract:
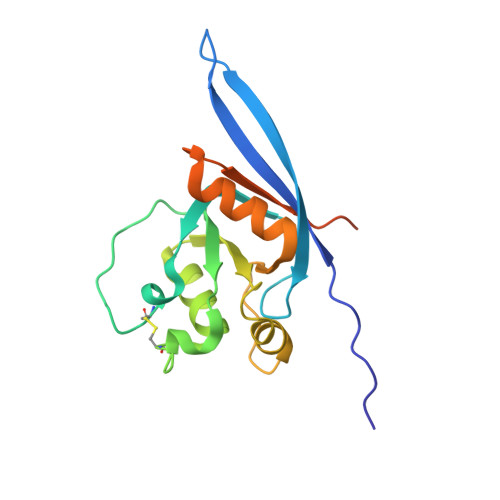
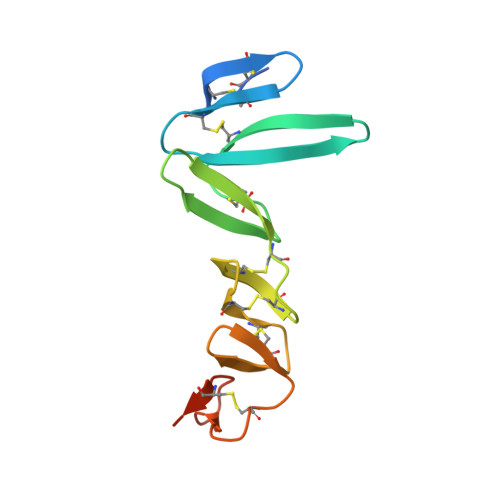
The four R-spondin (Rspo) proteins are secreted agonists of Wnt signalling in vertebrates, functioning in embryogenesis and adult stem cell biology. Through ubiquitination and degradation of Wnt receptors, the transmembrane E3 ubiquitin ligase ZNRF3 and related RNF43 antagonize Wnt signalling. Rspo ligands have been reported to inhibit the ligase activity through direct interaction with ZNRF3 and RNF43. Here we report multiple crystal structures of the ZNRF3 ectodomain (ZNRF3(ecto)), a signalling-competent Furin1-Furin2 (Fu1-Fu2) fragment of Rspo2 (Rspo2(Fu1-Fu2)), and Rspo2(Fu1-Fu2) in complex with ZNRF3(ecto), or RNF43(ecto). A prominent loop in Fu1 clamps into equivalent grooves in the ZNRF3(ecto) and RNF43(ecto) surface. Rspo binding enhances dimerization of ZNRF3(ecto) but not of RNF43(ecto). Comparison of the four Rspo proteins, mutants and chimeras in biophysical and cellular assays shows that their signalling potency depends on their ability to recruit ZNRF3 or RNF43 via Fu1 into a complex with LGR receptors, which interact with Rspo via Fu2.
Organizational Affiliation:
Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford OX3 7BN, UK.