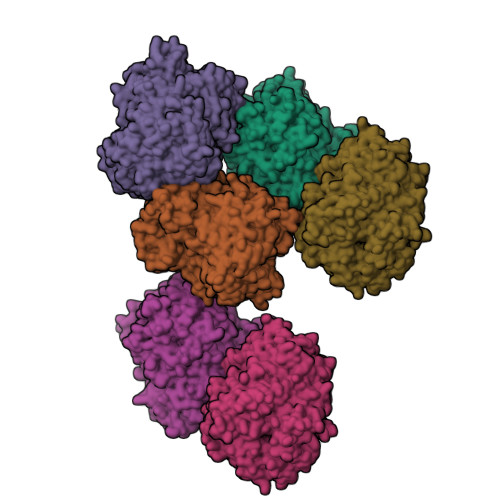
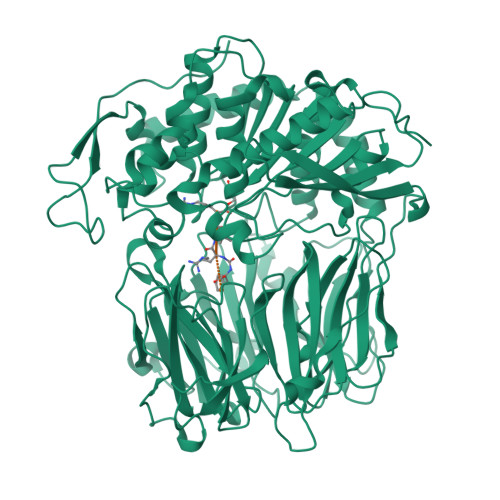
Crystal Structures of Trypanosoma Brucei Oligopeptidase B Broaden the Paradigm of Catalytic Regulation in Prolyl Oligopeptidase Family Enzymes.
Canning, P., Rea, D., Morty, R.E., Fulop, V.(2013) PLoS One 8: 79349
- PubMed: 24265767
- DOI: https://doi.org/10.1371/journal.pone.0079349
- Primary Citation of Related Structures:
4BP8, 4BP9 - PubMed Abstract:
Oligopeptidase B cleaves after basic amino acids in peptides up to 30 residues. As a virulence factor in bacteria and trypanosomatid pathogens that is absent in higher eukaryotes, this is a promising drug target. Here we present ligand-free open state and inhibitor-bound closed state crystal structures of oligopeptidase B from Trypanosoma brucei, the causative agent of African sleeping sickness. These (and related) structures show the importance of structural dynamics, governed by a fine enthalpic and entropic balance, in substrate size selectivity and catalysis. Peptides over 30 residues cannot fit the enzyme cavity, preventing the complete domain closure required for a key propeller Asp/Glu to fix the catalytic His and Arg in the catalytically competent conformation. This size exclusion mechanism protects larger peptides and proteins from degradation. Similar bacterial prolyl endopeptidase and archael acylaminoacyl peptidase structures demonstrate this mechanism is conserved among oligopeptidase family enzymes across all three domains of life.
Organizational Affiliation:
School of Life Sciences, University of Warwick, Coventry, United Kingdom.