Structural and Functional Perturbation of Giardia Lamblia Triosephosphate Isomerase by Modification of a Non-Catalytic, Non-Conserved Region.
Hernandez-Alcantara, G., Torres-Larios, A., Enriquez-Flores, S., Garcia-Torres, I., Castillo-Villanueva, A., Mendez, S.T., De La Mora-De La Mora, I., Gomez-Manzo, S., Torres-Arroyo, A., Lopez-Velazquez, G., Reyes-Vivas, H., Oria-Hernandez, J.(2013) PLoS One 8: 69031
- PubMed: 23894402
- DOI: https://doi.org/10.1371/journal.pone.0069031
- Primary Citation of Related Structures:
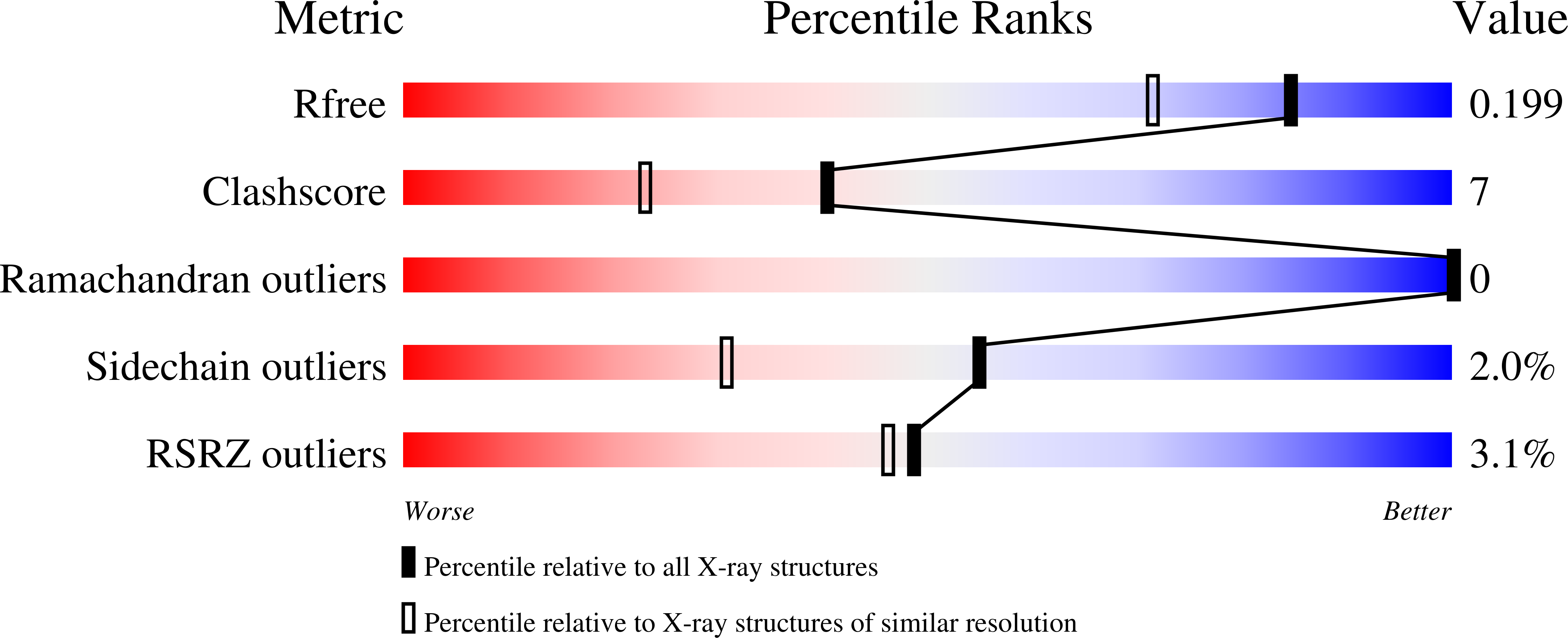
4BI5, 4BI6, 4BI7 - PubMed Abstract:
We have previously proposed triosephosphate isomerase of Giardia lamblia (GlTIM) as a target for rational drug design against giardiasis, one of the most common parasitic infections in humans. Since the enzyme exists in the parasite and the host, selective inhibition is a major challenge because essential regions that could be considered molecular targets are highly conserved. Previous biochemical evidence showed that chemical modification of the non-conserved non-catalytic cysteine 222 (C222) inactivates specifically GlTIM. The inactivation correlates with the physicochemical properties of the modifying agent: addition of a non-polar, small chemical group at C222 reduces the enzyme activity by one half, whereas negatively charged, large chemical groups cause full inactivation. In this work we used mutagenesis to extend our understanding of the functional and structural effects triggered by modification of C222. To this end, six GlTIM C222 mutants with side chains having diverse physicochemical characteristics were characterized. We found that the polarity, charge and volume of the side chain in the mutant amino acid differentially alter the activity, the affinity, the stability and the structure of the enzyme. The data show that mutagenesis of C222 mimics the effects of chemical modification. The crystallographic structure of C222D GlTIM shows the disruptive effects of introducing a negative charge at position 222: the mutation perturbs loop 7, a region of the enzyme whose interactions with the catalytic loop 6 are essential for TIM stability, ligand binding and catalysis. The amino acid sequence of TIM in phylogenetic diverse groups indicates that C222 and its surrounding residues are poorly conserved, supporting the proposal that this region is a good target for specific drug design. The results demonstrate that it is possible to inhibit species-specifically a ubiquitous, structurally highly conserved enzyme by modification of a non-conserved, non-catalytic residue through long-range perturbation of essential regions.
Organizational Affiliation:
Laboratorio de Bioquímica-Genética, Instituto Nacional de Pediatría, Secretaría de Salud, Mexico City, Mexico.