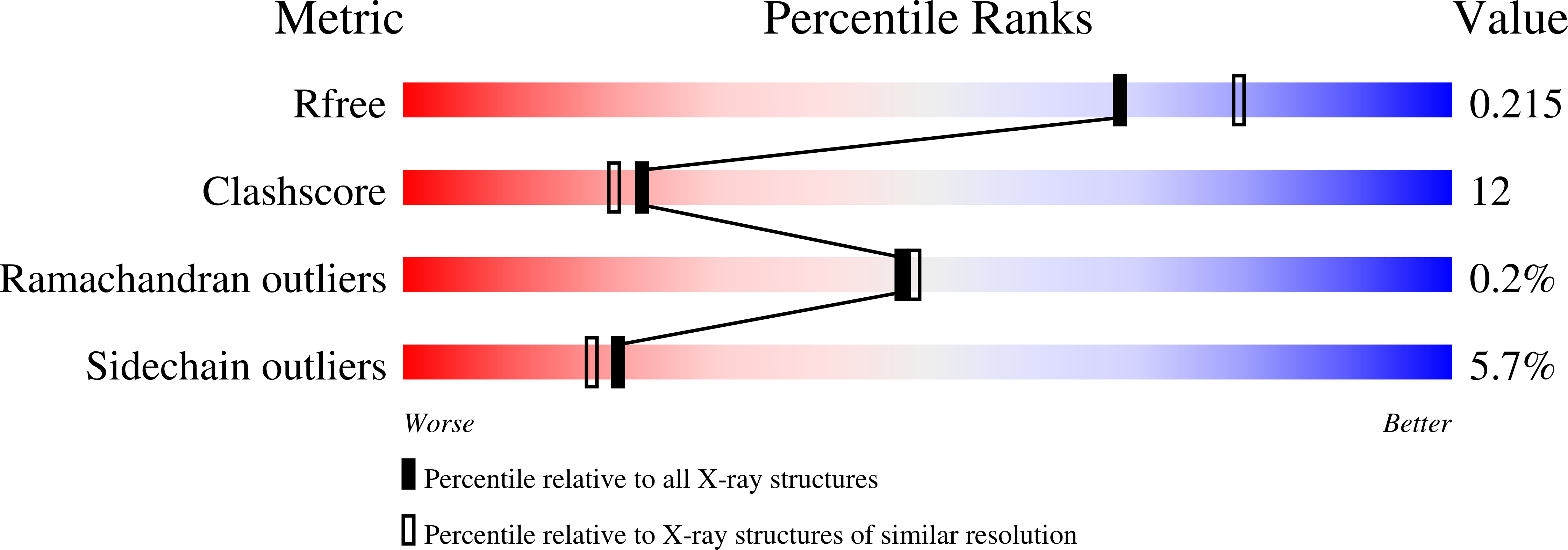Gfp-Like Phototransformation Mechanisms in the Cytotoxic Fluorescent Protein Killerred Unraveled by Structural and Spectroscopic Investigations.
De Rosny, E., Carpentier, P.(2012) J Am Chem Soc 134: 18015
- PubMed: 23025285
- DOI: https://doi.org/10.1021/ja3073337
- Primary Citation of Related Structures:
4B30 - PubMed Abstract:
KillerRed (KR) is a red fluorescent protein recognized as an efficient genetically encoded photosensitizer. KR generates reactive oxygen species via a complex process of photoreactions, ending up in photobleaching, the mechanism of which remains obscure. In order to clarify these mechanisms, we focus on a single mutant V44A (A44-KR) exhibiting the solely green component of KR. We report on the laser-induced structural transformations of A44-KR at cryogenic temperature, which we have investigated by combining UV-vis fluorescence/absorption spectroscopy with X-ray crystallography. Like the well-known GFP, A44-KR possesses a mixture of protonated (A) absorbing at 397 and deprotonated (B) absorbing at 515 nm chromophores, which are stressed by intense prolonged violet and blue laser sources. Both illuminations directly drive the B-chromophores toward a bleached trans isomerized form. A-type chromophores are sensitive only to violet illumination and are phototransformed either into a deprotonated green fluorescent form by decarboxylation of E218 or into a bleached form with a disordered p-hydroxybenzylidene. In crystallo spectroscopy at cryo-temperature allowed the identification and dissection of an exhaustive scheme of intermediates and end-products resulting from the phototransformation of A44-KR. This constitutes a framework for understanding the photochemistry of the photosensitizer KillerRed.
Organizational Affiliation:
Institut de Biologie Structurale Jean-Pierre Ebel, Groupe Métalloprotéines, UMR 5075, Université Joseph Fourier Grenoble 1, CEA, CNRS, 41 rue Horowitz, 38027, Grenoble Cedex 1, France.