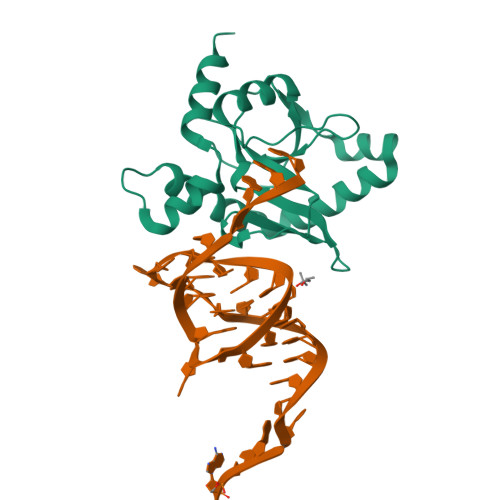
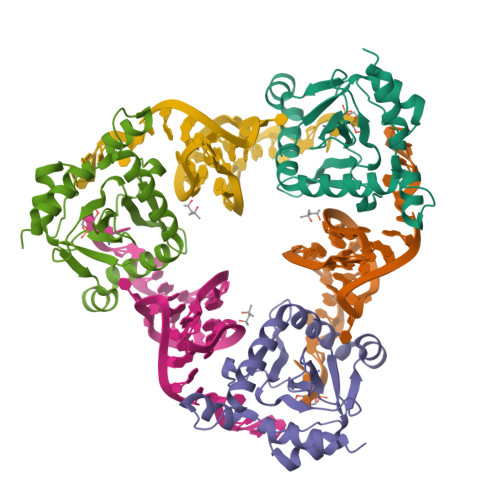
Selectivity and Self-Assembly in the Control of a Bacterial Toxin by an Antitoxic Noncoding RNA Pseudoknot.
Short, F.L., Pei, X.Y., Blower, T.R., Ong, S.L., Fineran, P.C., Luisi, B.F., Salmond, G.P.C.(2013) Proc Natl Acad Sci U S A 110: E241
- PubMed: 23267117
- DOI: https://doi.org/10.1073/pnas.1216039110
- Primary Citation of Related Structures:
4ATO - PubMed Abstract:
Bacterial small RNAs perform numerous regulatory roles, including acting as antitoxic components in toxin-antitoxin systems. In type III toxin-antitoxin systems, small processed RNAs directly antagonize their toxin protein partners, and in the systems characterized the toxin and antitoxin components together form a trimeric assembly. In the present study, we sought to define how the RNA antitoxin, ToxI, inhibits its potentially lethal protein partner, ToxN. We show through cross-inhibition experiments with the ToxIN systems from Pectobacterium atrosepticum (ToxIN(Pa)) and Bacillus thuringiensis (ToxIN(Bt)) that ToxI RNAs are highly selective enzyme inhibitors. Both systems have an "addictive" plasmid maintenance phenotype. We demonstrate that ToxI(Pa) can inhibit ToxN(Pa) in vitro both in its processed form and as a repetitive precursor RNA, and this inhibition is linked to the self-assembly of the trimeric complex. Inhibition and self-assembly are both mediated entirely by the ToxI(Pa) RNA, with no requirement for cellular factors or exogenous energy. Finally, we explain the origins of ToxI antitoxin selectivity through our crystal structure of the ToxIN(Bt) complex. Our results show how a processed RNA pseudoknot can inhibit a deleterious protein with exquisite molecular specificity and how these self-contained and addictive RNA-protein pairs can confer different adaptive benefits in their bacterial hosts.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, Cambridge CB2 1QW, United Kingdom.